Detection method and primers for evaluating enrichment result of porcine histone modification
A detection method and histone technology, applied in the field of ChIP-seq, can solve the problems of high cost of ChIP-seq, poor stability between materials, few applications, etc., and achieve the effect of saving time and economic cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
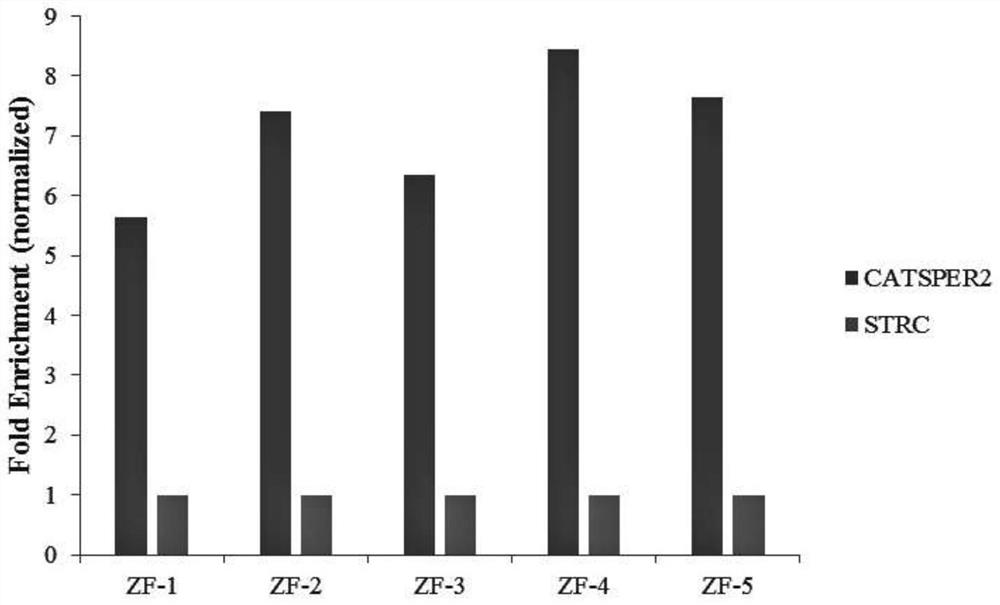
[0067] Example 1, quantitative detection of H3K4ME3 and H3K27AC enrichment in porcine adipose tissue.
[0068] In this embodiment, pig adipose tissue is used as a sample, and its specific operation steps are as follows,
[0069] 1) First take a 50ml centrifuge tube, add 50ml PBS 100ul PIS 200ul PMSF to pre-cool on ice, and then sample 0.5g of muscle tissue.
[0070] 2) Cut the sample into small pieces with scissors, add PBS to make it 30ml, then add 840ul 37% formaldehyde, and shake gently on a shaker at room temperature for 15min.
[0071] 3) Add 3.42ml of 2.0M glycine to make the final concentration 0.2M, shake gently on a shaker at room temperature for 5min.
[0072] 4) Centrifuge at room temperature for 5 minutes at 2500 rpm.
[0073] 5) The lower layer was washed twice with PBS, and centrifuged at 2500 rpm at 4° C. for 5 min.
[0074] 6) The liquid was discarded, and the precipitate was homogenized with cell lysate; wherein, the amount of cell lysate added was 3 mL of ...
Embodiment 2
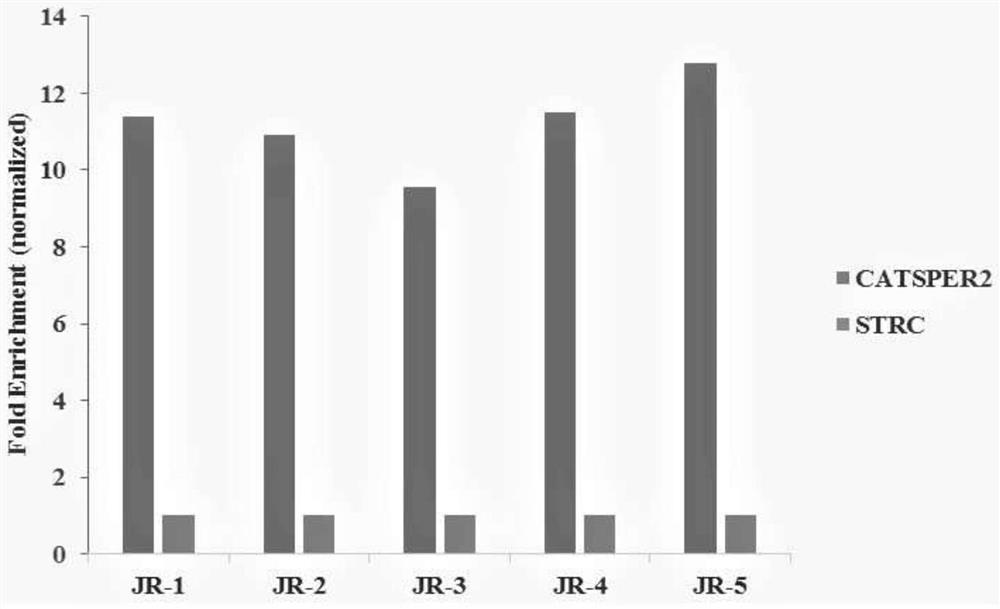
[0092] Example 2, quantitative detection of H3K4ME3 and H3K27AC enrichment in porcine muscle tissue.
[0093] In this embodiment, pig muscle tissue is used as a sample, and the specific operation steps are the same as those in Embodiment 1.
[0094] The result is as figure 2 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com