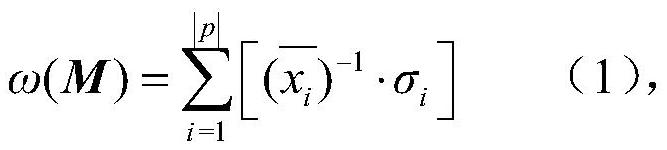Method of identifying cancer drive pathway
A cancer and pathway technology, applied in instrumentation, genomics, proteomics, etc., can solve problems such as low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048]A method for identifying cancer-driven pathways includes the following steps:
[0049]1) Construct a weighted non-binary mutation matrix:
[0050]Existing glioblastoma GBM somatic mutation matrixCopy number variation matrixAnd gene expression matrixMedium mutation matrixCopy number variation matrixAnd gene expression matrixThe middle row represents the same sample set p of a cancer, and the columns represent the gene set GS, GCAnd GE, In the matrixMiddle, sij ∈{0,1}(i=1,2,...,|p|,j=1,2,...,|GS|), j gene mutation in i sample, sij The value is 1, otherwise the value is 0; matrixEach element in cij ∈{-2,-1,0,1,2}(i=1,2,...,|p|,j=1,2,...,|GC|), represents the copy number variation value of j gene in i sample; in the matrixMiddle eij ∈R(i=1, 2,..., |p|, j=1, 2,..., |GE|), represents the expression of j gene in i sample; let the matrixGene set GA=GS∪GC, The sample set is p, letaij ∈{0,1}(i=1,2,...,|p|,j=1,2,...,|GA|), whereIs the mutation matrix, when sij The value is 1 or when the j gene...
Embodiment 2
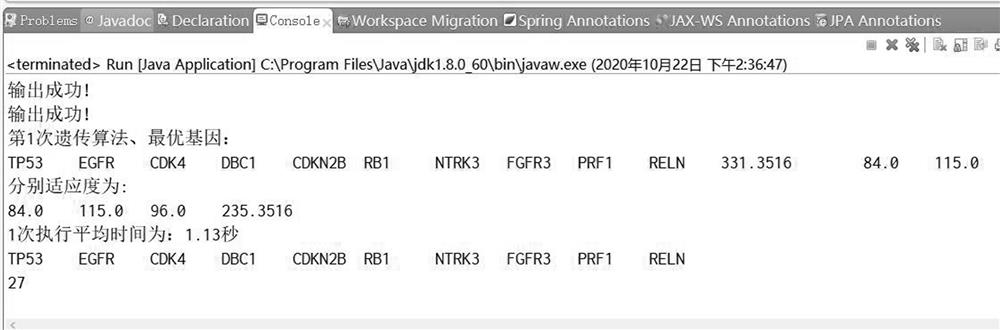
[0079]Set λ in step 1) of this example1= 3 and λ2=7, construct a weighted non-binary mutation matrix A|p|×|G| , Where |p|=90, |G|=920;
[0080]Step 7 of this example) Enter the weighted non-binary mutation matrix A|p|×|G| , Where |p|=90, |G|=920, the model in formula (2), the drive path size k=10, CGA-MWS related parameters, population size P=460, mutation probability Pm=0.3, the maximum evolutionary algebra maxstep=1000, the optimal value is kept constant threshold maxt=10;
[0081]In this example, step 9) get the drive path with size k=10, the operation diagram is as followsfigure 2 Shown.
[0082]The remaining steps are the same as in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com