In vivo homology directed repair in heart, skeletal muscle, and muscle stem cells
A heart cell, muscle technology, applied in the field of in vivo homology-directed repair in heart, skeletal muscle and muscle stem cells, can solve the problems of untested multi-organ HDR feasibility and inefficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
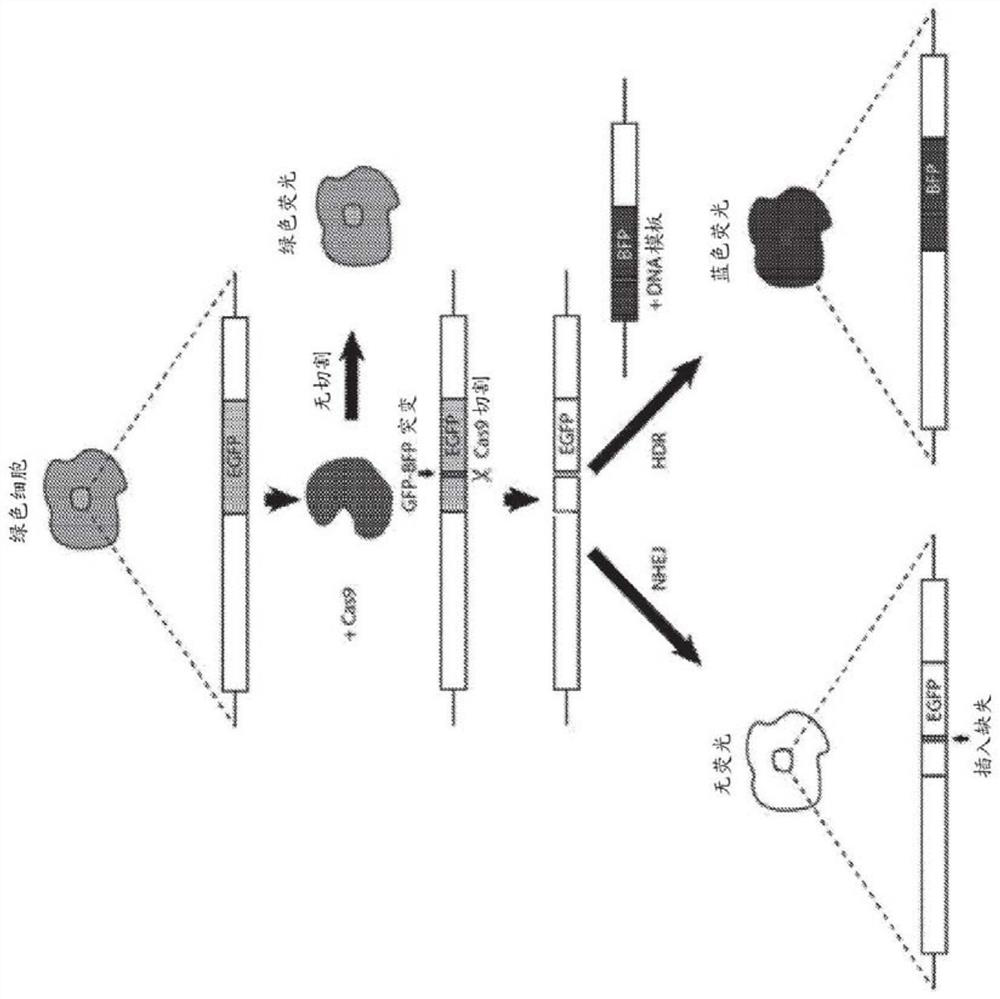
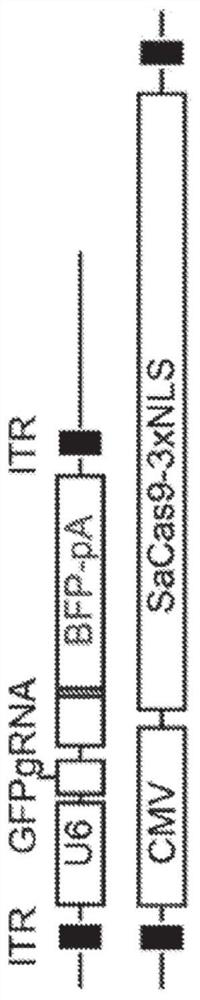
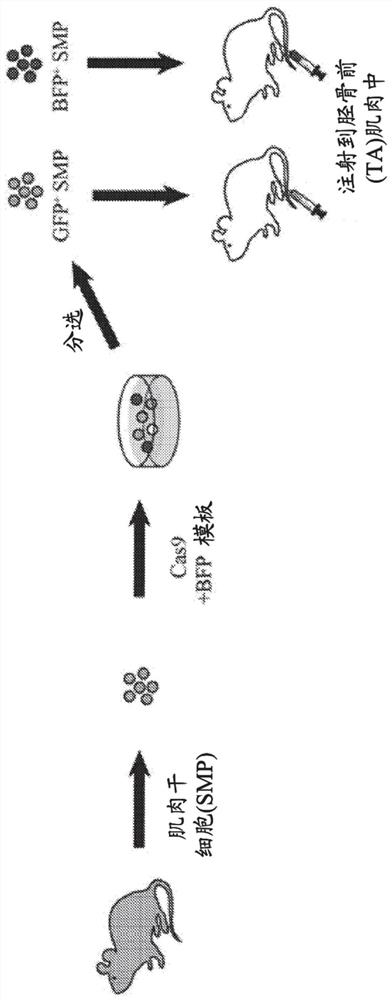
[0113] To sensitively detect in vivo gene editing events via CRISPR / Cas9, a strongly enhanced green fluorescent protein (GFP) signal using ubiquitous expression was developed 8 A fluorescent protein-based reporter system for transgenic mouse strains ( Figure 1A ). Based on published BFP variants 9-11 The blue fluorescent protein (BFP) sequence was designed to carry a minimum of 2 base substitutions (C197G and T199C) compared to the GFP sequence. This simple modification allows easy differentiation of two fluorescent proteins by fluorescence-activated cell sorting (FACS) ( Figure 5A-Figure 5D ). The same 2 base substitutions also created a Btgl site for restriction fragment length polymorphism (RFLP) analysis. A single guide RNA (sgRNA) targeting a substitution site in GFP was designed to be compatible with the Cas9 protein (SaCas9) from S. + / - ; Efficient disruption of GFP signaling was tested in tail tip fibroblasts (TTF) of mdx mice ( Figure 5B , Figure 5C ). Thi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com