A method, device and storage medium for diploid assembly
A diploid and haplotype technology, applied in the fields of genomics, proteomics, instruments, etc., can solve the problems of limited application prospects, typing impact, and high genomic heterozygosity, and achieve the effect of overcoming difficulties in obtaining
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
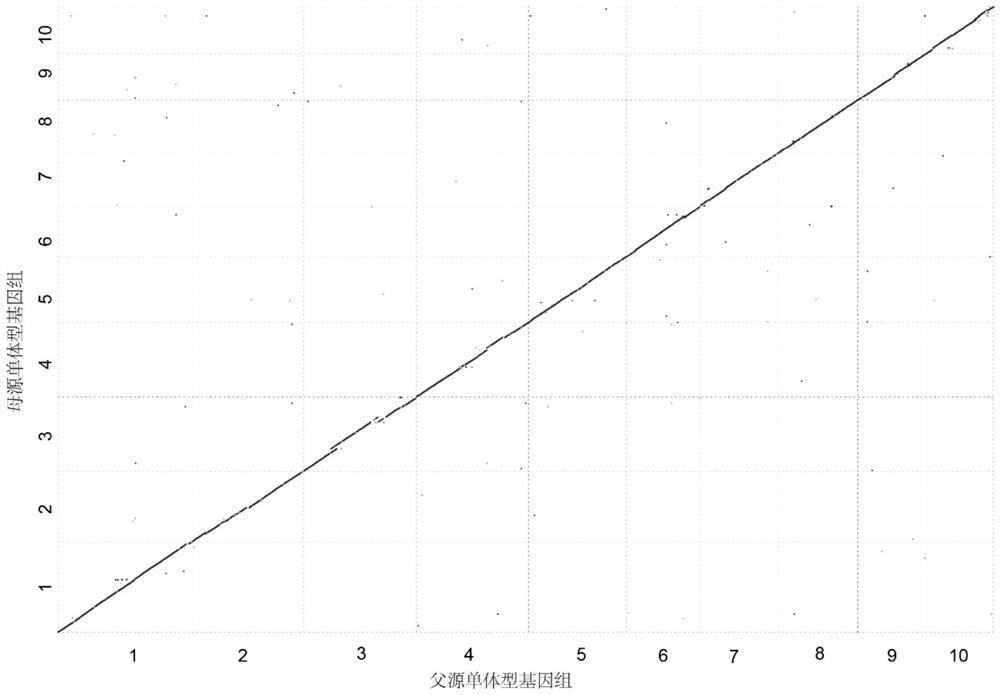
[0085] In this case, a diploid assembly test was performed on the F1 generation high-heterozygous genome of hybrid maize. The parents of the hybrid maize were maize B73 and maize SK, respectively. The diploid genome size is about 4.5Gb. In this experiment, this example was based on PacBio HiFi data, Hi-C data, and gamete single-cell data to perform diploid assembly of this hybrid F1 individual. details as follows:
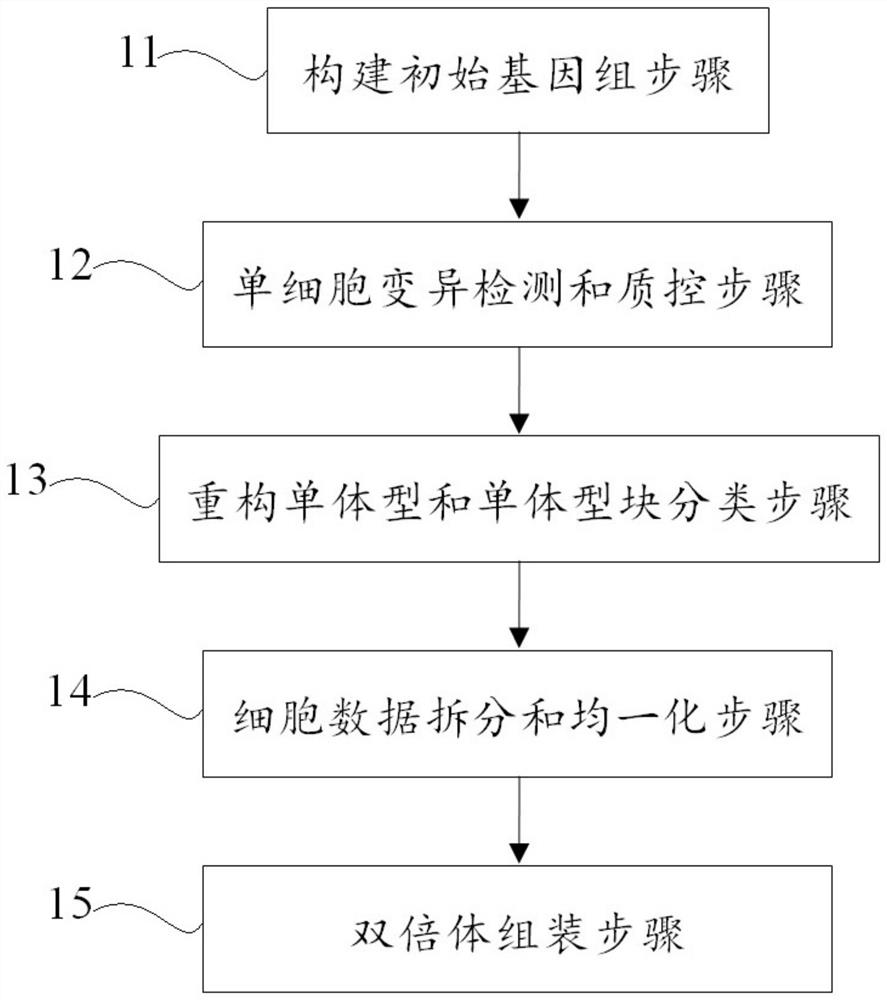
[0086] 1. Construction of the initial genome
[0087] In this example, the PacBio HiFi data of hybrid F1 individuals were obtained based on the parental genome sequence simulation. The genomic data of the parents are published public data.
[0088] The SK genome download address in this example is:
[0089] https: / / db.cngb.org / search / assembly / CNA0002536 / ;
[0090] The B73 genome download address is:
[0091] ftp: / / ftp.ensemblgenomes.org / pub / plants / release-50 / fasta / zea_mays / dna / ;
[0092] The download address of pbsim software is: https: / / github.com / pfaucon / P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com