Inductive base editing system and application thereof
A base editing and inducible technology, applied in the field of genetic engineering, can solve problems such as amplification of off-target consequences, and achieve the effect of shortening editing time and reducing off-target editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1 constructs split-A3A plasmid
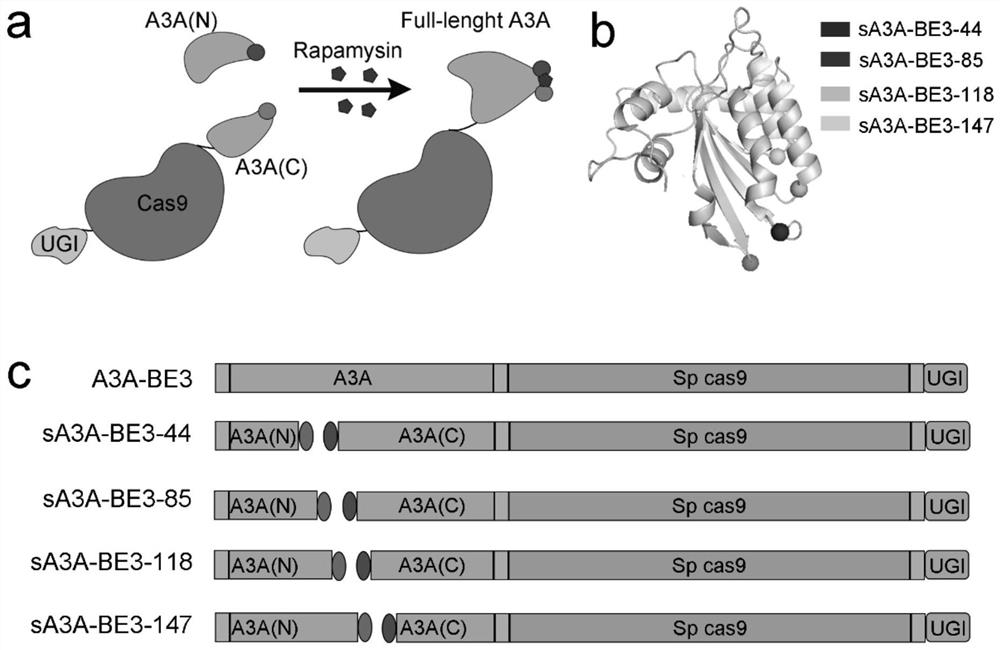
[0033] The split-A3A plasmid was constructed on the basis of A3A-BE3 to form a series of N-terminal split-A3A and its corresponding C-terminal split-BE3-A3As ( figure 1 b). Such as figure 1 As shown in c, four groups of split-A3A-BE3 (sA3A-BE3) were constructed and named after their amino acid splitting sites, namely sA3A-BE3-44, sA3A-BE3-85, sA3A-BE3-118 and sA3A- BE3 is -147 respectively.
[0034] figure 1 It is the design of inducible split-A3A-BE3; among them, panel a shows the schematic diagram of the reassembly of inducible split-A3A-BE3, and rapamycin induces the dimerization of FRB and FKBP fused to the N-terminus and C-terminus of A3A, respectively To form a heterodimer, thereby completing the assembly of a functional A3A-BE3 base editor. Figure b is a schematic diagram of the structure of A3A (PDB: 5keg). The four unstructured loops opposite the ssDNA-A3A binding interface are candidate cleavage sites, represen...
Embodiment 2
[0035] Example 2 split-A3A base editing
[0036] 1. Detection of inducible base editing activity of sA3A-BE3
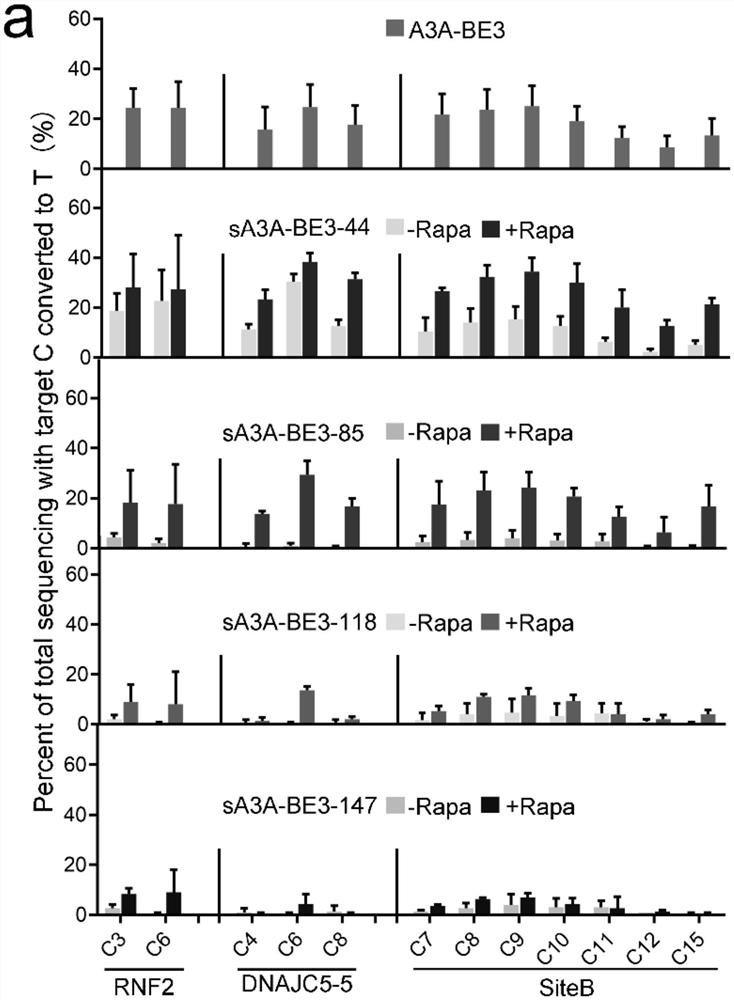
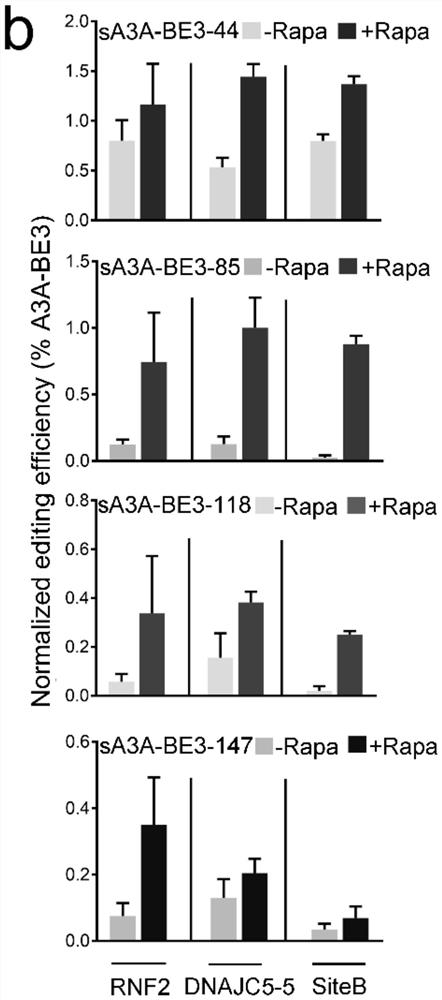
[0037] Each pair of split-A3A-BE3 editors was separately co-transfected with sgRNAs for each of the 3 endogenous targets into HEK293T cells. Such as figure 2 In a, all sA3A-BE3s exhibit inducible editing activity, albeit with different potentials. Among these sA3A-BE3, sA3A-BE3-44 edited most efficiently at all 3 target sites when induced with 200 nM rapamycin, a concentration frequently used in the split system of FRB / FRBP. However, its activity remained essentially unchanged under uninduced conditions. On average, uninduced sA3A-BE3-44 exhibited comparable activity to full-length A3A-BE3 ( figure 2 b). Transfection of the C-terminal portion of sA3A-BE3-44 did not produce any detectable editing, suggesting that the background editing activity of sA3A-BE3-44 was not due to the residual deamination activity of C-terminal A3A, but probably due to the autologous d...
Embodiment 3
[0042] Further characterization of embodiment 3 split-A3A
[0043] To examine whether the cleavage of A3A at amino acid 85 changes the base editing mode of A3A-BE3, including editing window, sequence preference and editing purity of C→T.
[0044] To determine the editing window, 3 targets with multiple cytosines within the editing window were selected ( Figure 4 a). Such as Figure 4 a, compared with full-length A3A-BE3, sA3A-BE3-85 does not significantly shift the position of the editing window or affect the width of the editing window. We then tested whether sA3A-BE3-85 has a different sequence preference than full-length A3A-BE3, as previous studies have shown that the unstructured loop between deaminase helices plays an important role in substrate recognition (Salter et al. 2016). The nine targets were divided into four groups according to the NC motif (GC, CC, AC and TC), and the sequence preference of sA3A-BE3-85 and full-length A3A-BE3 was compared ( Figure 4 b)....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com