Deep learning-based chromosome uploidy prediction system and method, terminal and medium
A technology of deep learning and prediction methods, applied in the field of biomedicine, can solve the problems of long discrimination cycle, low penetration rate, high cost, etc., and achieve the effect of guaranteeing training and testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
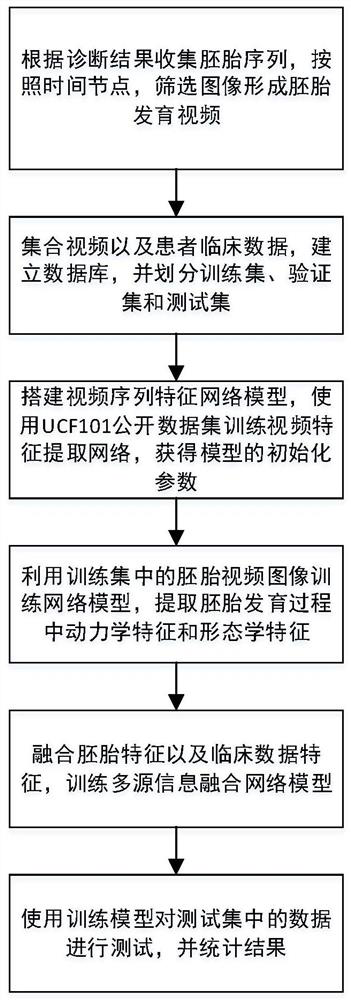
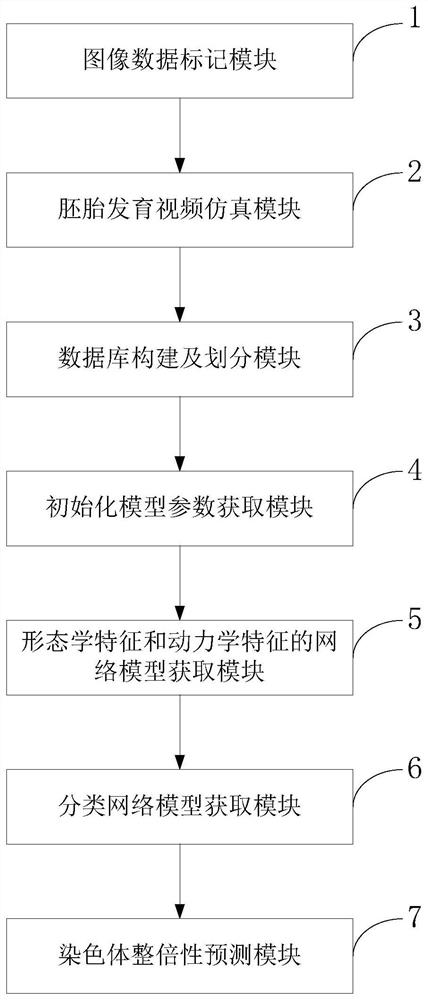
[0090] Collect embryo sequences with third-generation IVF diagnostic results, and label image files in sequence according to the temporal sequence of embryo development. Combined with the consensus of domestic and foreign embryologists and the observation experience of embryologists, the images of embryos in different scoring stages such as pronuclear stage, cleavage stage, blastocyst stage, etc. are selected to form a representative embryonic development video, which replaces the complete embryonic development video. embryonic development process.
[0091] The basic clinical data of the patient to which the embryo belongs is collected and integrated with the embryo video data to establish a database with chromosomal euploidy diagnosis results. Divide the data in the database into training set, validation set and test set according to the ratio of 8:1:1;
[0092] Build a video sequence feature network model, use the UCF-101 natural video image data set to initialize and train...
Embodiment 2
[0109] 1. Data collection and preprocessing stage
[0110] The present invention collects a total of 896 embryos with diagnosis results from the reproductive center, and at the same time, collects the data recorded by the embryologist corresponding to the embryos and the clinical data of the patient. According to the consensus of experts at home and abroad and the time point when embryologists observe the embryo, the embryo images of different developmental stages such as prokaryotic stage, cleavage stage and blastocyst stage are selected from the complete embryo image and combined to form embryo development video. The data recorded by the embryologist and the clinical data of the patients were counted, the mean and variance were calculated, the values were normalized, and the missing items in the data were filled with 0. Integrate the embryo video and the normalized value into a complete data, take the diagnosis result of chromosome euploidy as the prediction label of the p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com