Nucleic acid molecule targeting CYP4V2 gene mutation site and application thereof
A nucleic acid molecule and gene technology, applied in the field of gRNA and donor nucleic acid molecules, can solve problems such as limited curative effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0109] Example 1 Design sgRNA
[0110] 1. Screening of sgRNA
[0111] According to the DNA sequence of the CYP4V2 gene, the PAM sequence in the intron region between exon 6 and exon 7 of CYP4V2 is designed as NNGRRT and NNGRR (Staphylococcus aureus, Staphylococcus aureus, SA; SaCas9), with a length of 21bp sgRNA.
[0112] According to the scores, a total of 7 sgRNAs (5 for NNGRRT and 2 for NNGRR) were designed, and their sequences are shown in Table 1.
[0113] Table 1. CYP4V2-HITI method benchling online design of sgRNA
[0114]
[0115]
[0116] 2. sgRNA synthesis
[0117] According to the restriction site Bbs1 sequence, add the Bbs1 restriction site in the upstream and downstream of the designed sgRNA; and design corresponding primers within 400 bp upstream and downstream of each sgRNA. The corresponding oligonucleotide sequences and primers were designed in Table 2.
[0118] Table 2 Design of sgRNA
[0119]
[0120]
[0121] 3. The specific steps of sgRNA...
Embodiment 2
[0232] Example 2 Verifying the impact of sgRNA targets on cleavage
[0233] 1. The four sgRNAs correspond to the pMD19-T minigene plasmid, and its control negative plasmid and positive plasmid design.
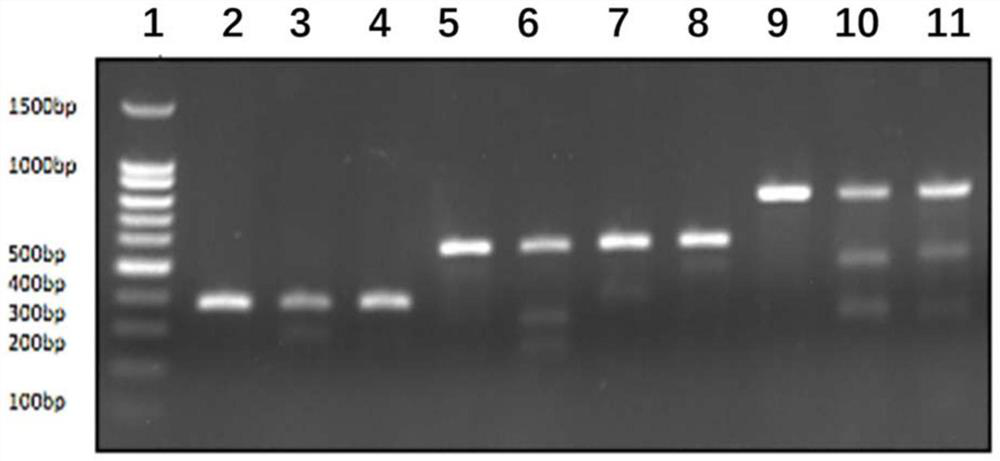
[0234] According to the donor HITI method, the characteristics of PAM+3bpsgRNA / 18bp sgRNA fragments will remain after insertion, and minigene1.2.3.4 is designed to correspond to sgRNA1.2.3.4. The minigene fragments are as follows Figure 4 shown.
[0235] Second, the minigene plasmid construction steps are as follows
[0236] 1. Synthesize minigene DNA fragments and positive control (normal wild-type intron, which does not affect cleavage) and negative control (patient mutated intron, which affects cleavage) DNA fragments.
[0237] 2. Restriction plasmid vector
[0238] The plasmid vector is PMD-19T-MCS, see the plasmid map Figure 5 .
[0239] (1) Digest the plasmid with KpnI enzyme and MluI
[0240] In CutSmart buffer, the KpnI enzyme and MluI enzyme were incubated with...
Embodiment 3
[0317] Embodiment 3 Design and screening of Donor
[0318] 1. Carrier construction:
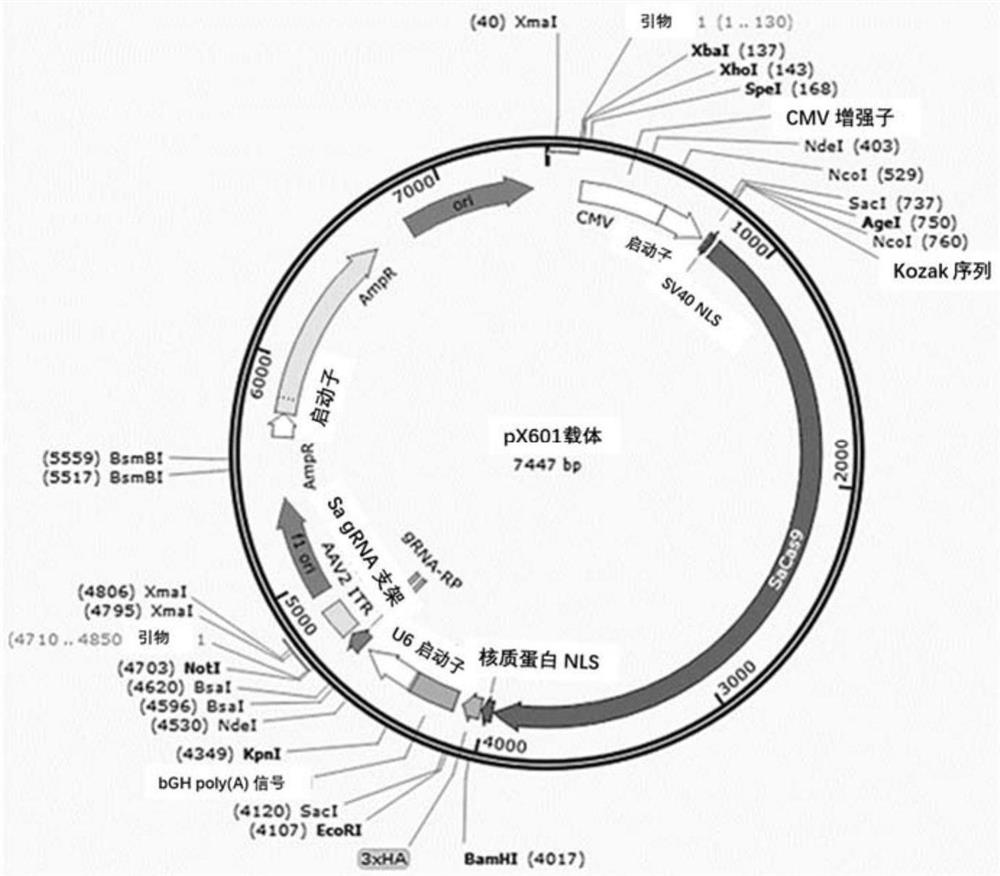
[0319] The sequence between intron 6 and exon 7-11 of the CYP4V2 wild-type gene was used as the Donor sequence; the Donor sequence and the EGFP reporter gene were constructed together into the pX601 vector (pX601-sgRNA1 to pX601-sgRNA4 of sgRNA1-4 carrier) to obtain the pX601-donor(1-4)-EGFP carrier; the sgRNA carrier uses the pX601-sgRNA(1-4) carrier described in Example 1. PX601 plasmid map as figure 1 shown.
[0320] Wherein, the length of intron 6 is adjusted according to the sgRNA cutting site, the Donor sequence of exons 7-11 is shown in SEQ ID NO:38; the EGFP sequence is shown in SEQ ID NO:39.
[0321] 2. Transfection of iPSCs by PEI method
[0322] 1. The specific steps are as follows:
[0323] Take the sgRNA1 group as an example:
[0324] (1) Take four 6-well plates, inoculate iPSC cells, and perform transfection when the cell density grows to 80%;
[0325] (2) The first group...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com