Efficient prediction method for association relationship between circRNA and disease
A technology of correlation and disease, applied in the field of bioinformatics, can solve the problems of reducing accuracy, time-consuming, and not considering different subtle semantic meanings, and achieve the effect of reducing workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
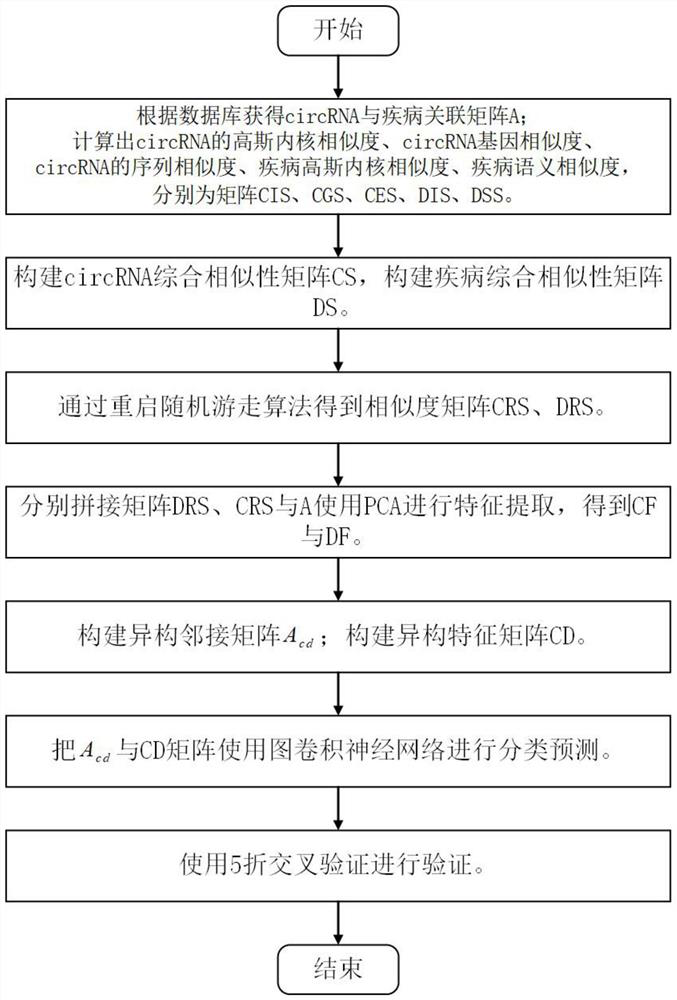
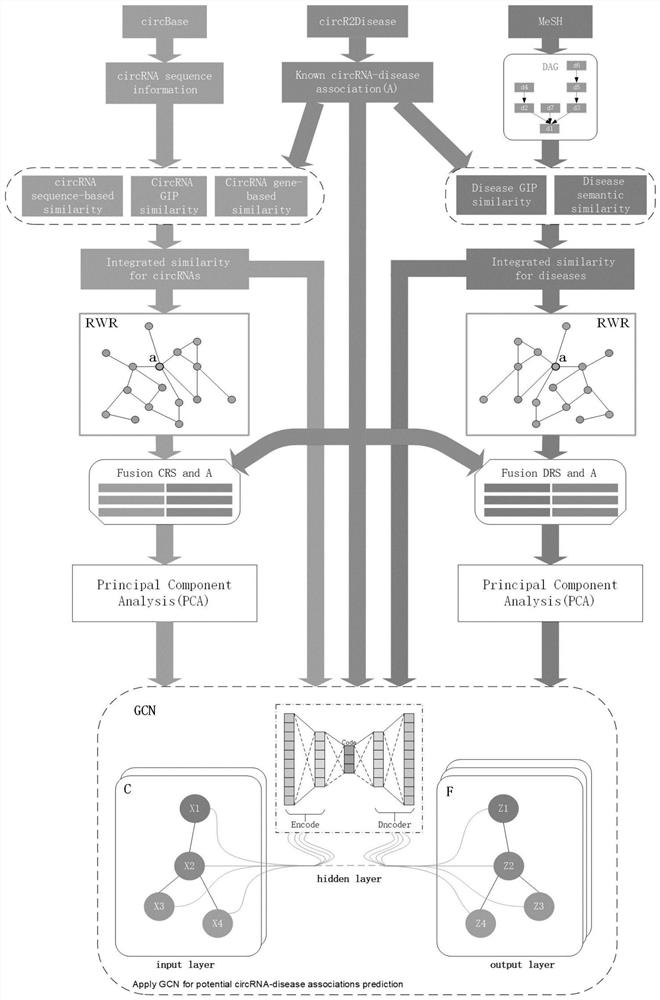
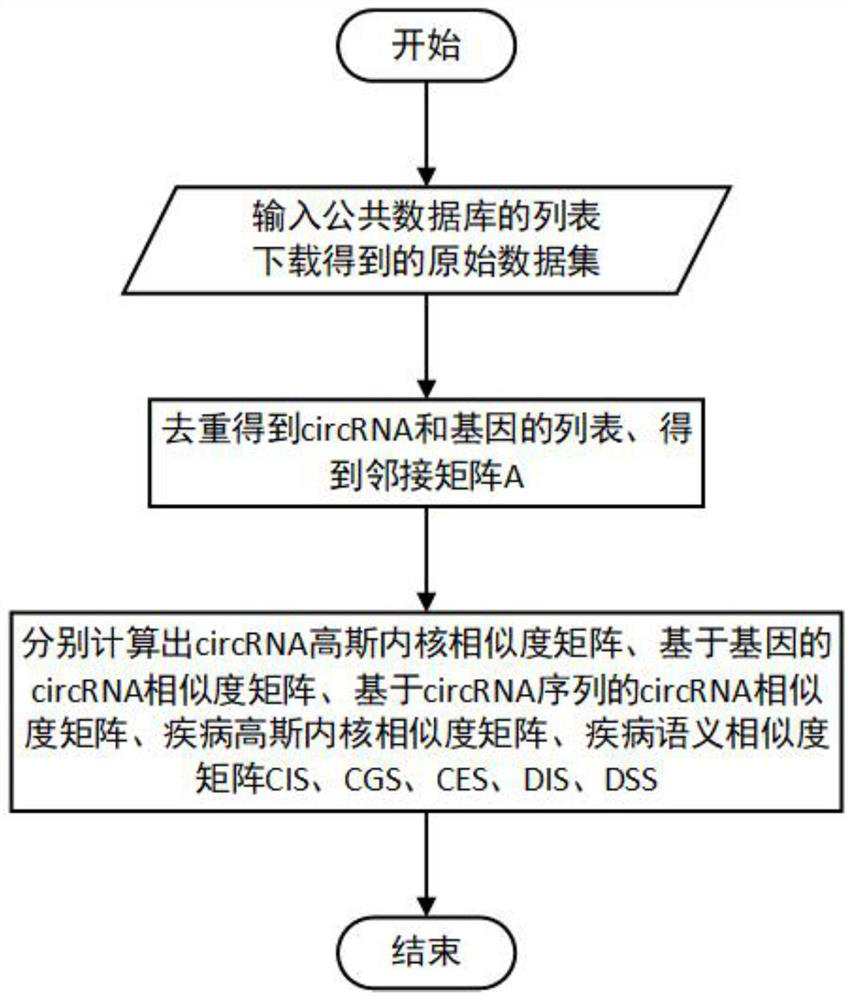
[0116] The present invention will be further described in detail below in conjunction with the embodiments and the accompanying drawings, but the embodiments of the present invention are not limited thereto. Example:
[0117] 1. Download the data set and download the circRNA-disease association data from the public database CircR2Disease. After deduplication, the total association between circRNA and disease is 48049. The data format is shown in the following list:
[0118] Table 1 circRNA list
[0119] Numbering circRNA name 1 hsa_circ_0000977 2 hsa_circ_0006220 3 hsa_circ_0001666 … … 533 Circ_ERC1
[0120] Table 2 List of diseases
[0121] Numbering disease name pmid 1 Pancreatic ductal adenocarcinoma 29255366 2 Primary great saphenous vein varicosities 29577902 3 Lung cancer 29550475 … … … 89 Atherosclerotic vascular disease 21151960
[0123] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com