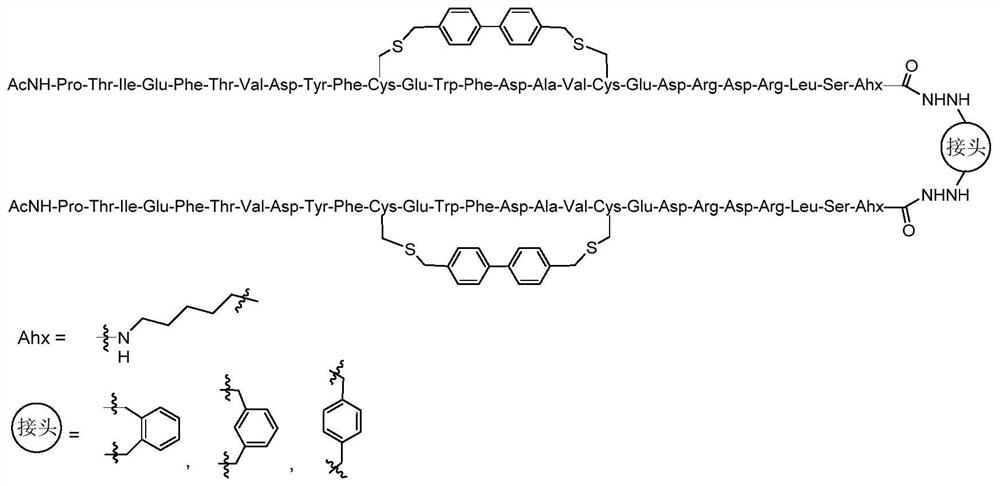
Cyclic peptide combined with RBD site of novel coronavirus as well as preparation method and application of cyclic peptide
A technology of cyclic peptides and viruses, which is applied in the field of cyclic peptides combined with the RBD site of the new coronavirus and its preparation and application, which can solve the problems of inability to inhibit the infection of the new coronavirus and the inability to bind
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1: synthetic hydrazine-2-Cl-Trt resin
[0038]Weigh 268 mg of 2-CI-Trt-Cl resin (loading capacity: 0.56 mmol / g, 150 umol) into a solid-phase synthesis tube, add 3 mL of DMF to swell for 20 minutes. The resin was sequentially washed three times with DCM and twice with DMF. Add 3.5 mL 5% hydrazine hydrate (125 μL hydrazine hydrate + 3.33 mL DMF), shake at room temperature for 30 min, discard the waste liquid, and repeat once. Add 2.5 mL of 20% MeOH / DMF (2 mL DMF + 0.5 mL MeOH) and shake at room temperature to block the reaction for 20 min, wash with DMF for 5 times to obtain hydrazine-2-Cl-Trt resin.
Embodiment 2
[0039] Example 2: Fmoc solid-phase synthesis of linear hydrazide peptides
[0040] The resin prepared in Example 1 was transferred to a solid-phase synthesis tube, and the DMF mixture of DIC / Oxyma / 6-aminocaproic acid (10eq DIC: 10eq Oxyma: 10eq 6-aminocaproic acid, 2mL DMF) was added, React at 55°C for 40 minutes. After the resin was washed with DMF, 2 mL of blocking reagent (acetic anhydride: 2,6-lutidine: DMF=5:6:89) was added to block unlinked amine groups. After 2 min, the resin was washed three times with DMF. Then add 20% piperidine in DMF, shake at room temperature for 8 minutes, and discard the reaction solution. Repeat the 20% piperidine treatment step. After cleaning the resin three times with DMF, add the DMF mixture of DIC / Oxyma / Fmoc-Ser(tBu)-OH (10eq DIC:10eq Oxyma:10eq Fmoc-Ser(tBu)-OH, 2mL DMF), React for 40 minutes. Next, repeat the above operation for amino acid condensation, and condense Fmoc-protected amino acids Leu, Arg, Asp, Arg, Asp, Glu, Cys, Val, ...
Embodiment 3
[0042] Example 3: Side chain cyclization of linear hydrazide peptides
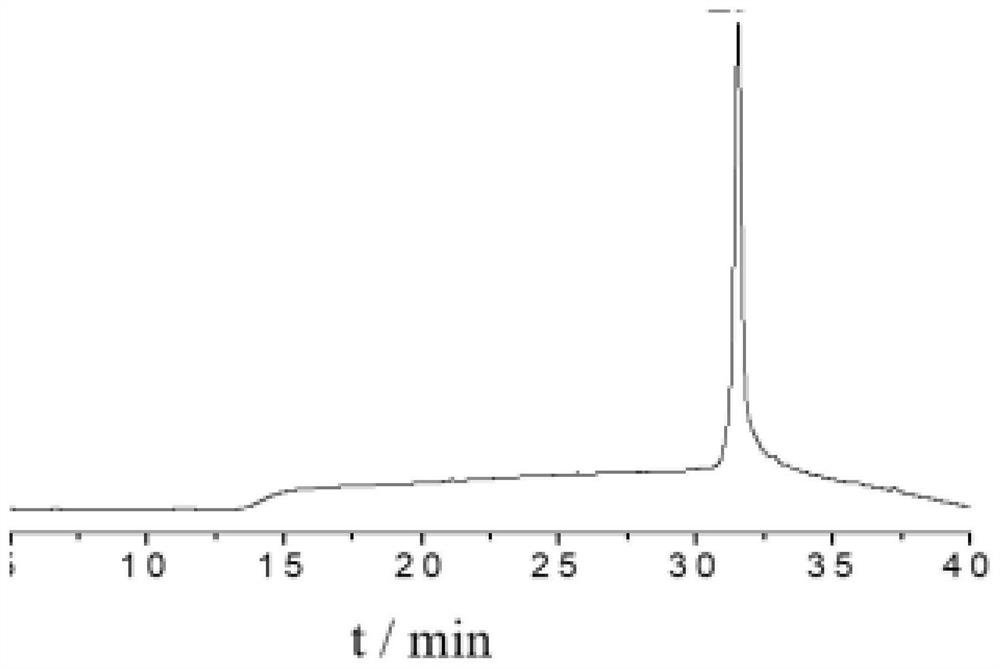
[0043] Dissolve 10 mg of linear peptide in 2.0 mL of DMF:H 2 O mixture (2:1, v:v), 1.4 mg of 4,4-dibromomethylbiphenyl was dissolved in 0.1 mL of DMF and added to the mixture with 1.0 M NH 4 HCO 3 The pH of the reaction mixture was adjusted to 8.0. After reacting at room temperature in a vibrating mixer for 2 h, add 3 mL of acetonitrile:H 2 O mixture (1:1, v:v, 0.1% TFA) to quench the reaction. After purification of the cyclized peptide by preparative HPLC and lyophilization, a white powder (~3 mg, 30%) was obtained as Figure 4 shown. The sequence of the side chain cyclized hydrazide polypeptide is as follows:
[0044]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com