ssr primer set for analysis of genetic structure of powdery mildew of rubber tree and its application
A technology of powdery mildew of rubber tree and population genetics, applied in the direction of proteomics, microbe-based methods, microorganisms, etc., can solve the problems of SSR marker research reports on powdery mildew of rubber tree, and achieve rich polymorphism, easy statistics, and expansion The effect of increasing stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 SSR primer development
[0027] (1) Genomic SSR locus analysis
[0028] Download the whole genome sequence of P. rubra from the NCBI (National Center for Biotechnology Information) website, and use MISA software to search for SSR sites of 2 to 6 nucleotides in the genome sequence, and the search condition is the number of repetitions of 2 nucleotides It must be ≥6, and the number of repetitions of 3-6 nucleotides must be ≥5.
[0029] A total of 2,720 SSR sites were found that met the search criteria, of which dinucleotide SSR sites were the most, with 1,619 sites; followed by tri-nucleotide SSR sites, with 682 sites; they accounted for 59.52 of the total SSR sites. and 25.07%; there were 330 tetranucleotide SSR sites, accounting for 12.13%; and neither pentanucleotide nor hexanucleotide SSR sites exceeded 50, accounting for less than 2%.
[0030] Table 1. Distribution of SSRs in the Genotypic SSR of Helicobacter
[0031]
[0032] (2) SSR primer design an...
Embodiment 2
[0053] Embodiment 2, utilizes SSR primer to carry out population genetic structure analysis to rubber tree powdery mildew in different regions
[0054] (1) 15 powdery mildew strains (Table 6) were obtained from Guangdong, Yunnan and Hainan respectively, and after DNA was extracted with the Genome Extraction Kit of Tiangen, PCR amplification was performed, and the specific operation was the same as that in Example 1.
[0055] Table 6. Information on 45 rubber tree powdery mildew strains
[0056]
[0057]
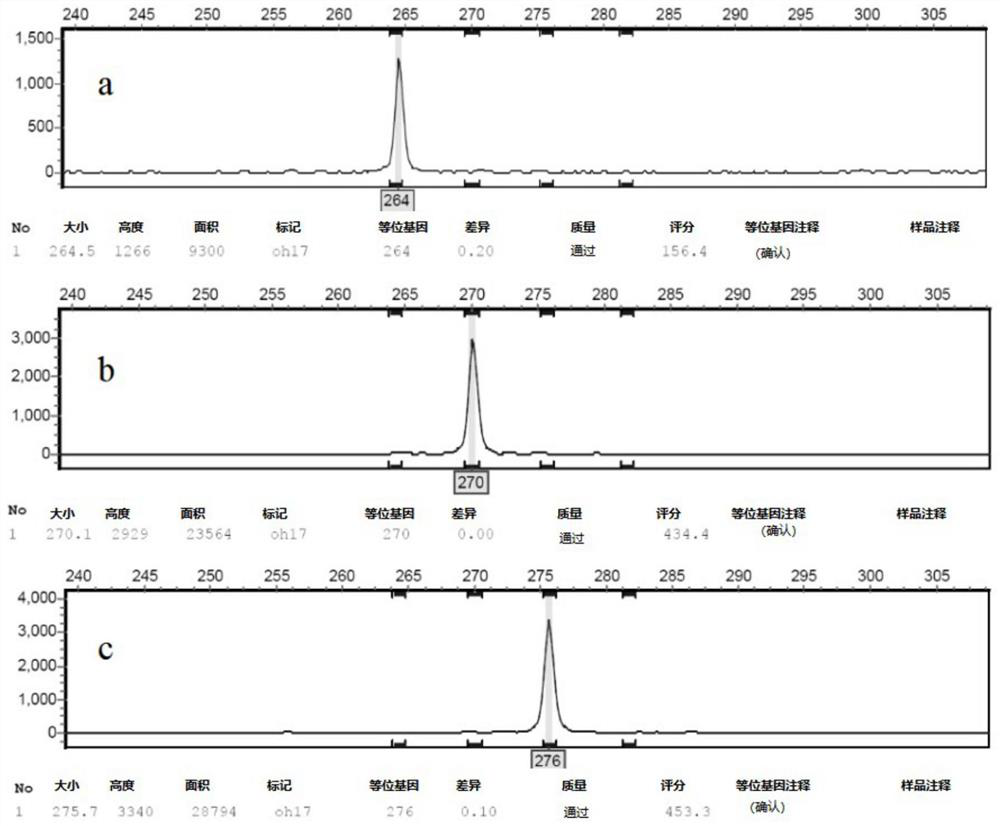
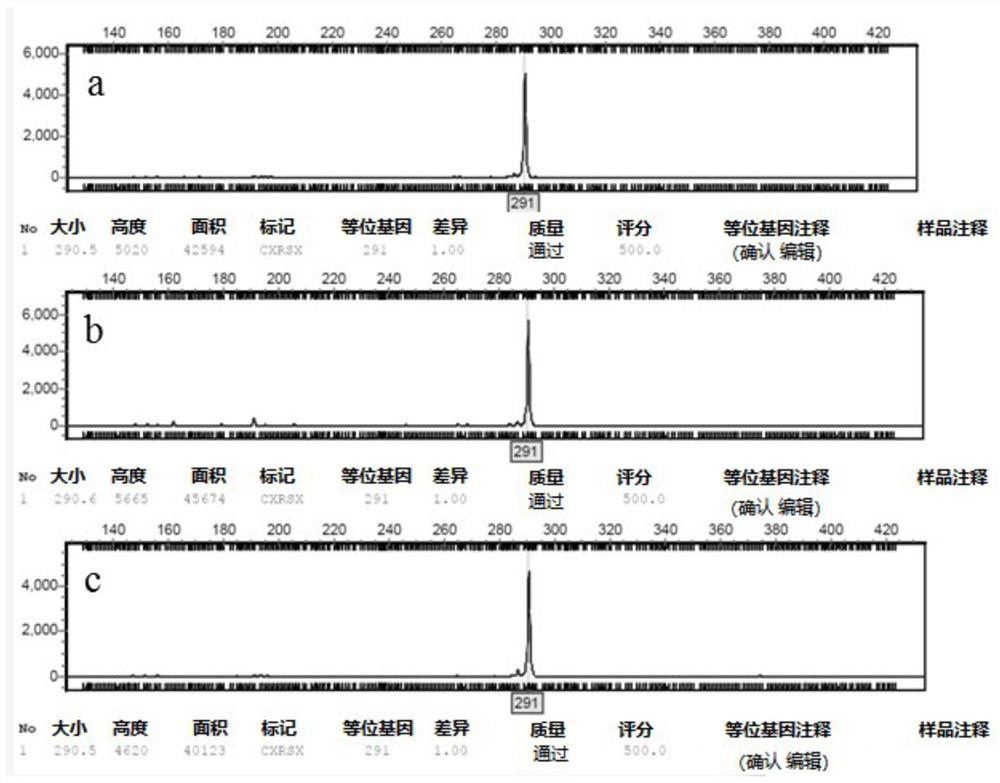
[0058] (2) Capillary fluorescence electrophoresis was used to detect the amplified products using DNA sequencer ABI3730x1, and GeneMarker V3.0.0 was used to read the capillary electrophoresis data, and the detection results were counted (Table 7).
[0059] Table 7. Amplification results of 16 pairs of SSR primers in 45 rubber tree powdery mildews
[0060]
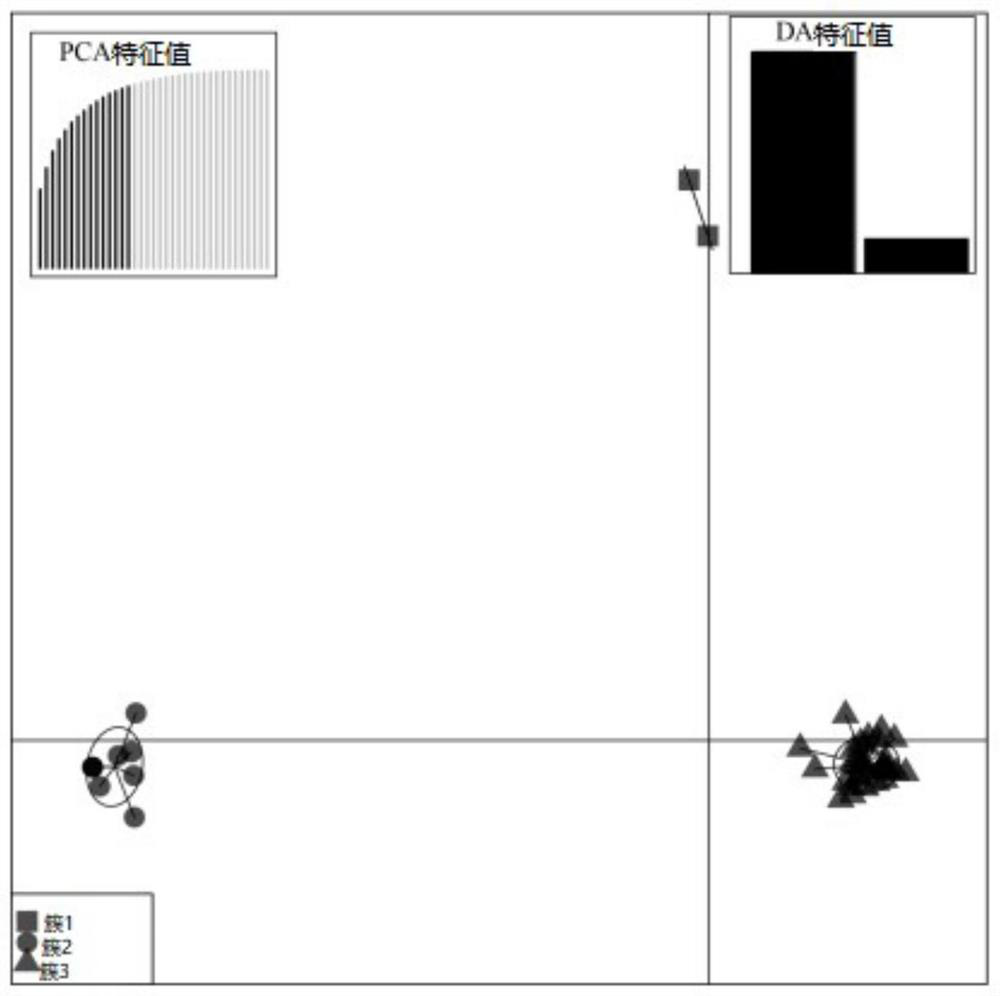
[0061] (3) GenAlEx 6.5 software was used to calculate the observed allele number Na, effective allele number N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com