A Molecular Marker Method for Phylogenetic Relationship and Cluster Analysis of Bactrocera citrus
A technology of Bactrocera citrus and kinship, applied in the direction of biochemical equipment and methods, measurement/inspection of microorganisms, etc., can solve the problems of inability to accurately and quickly detect and identify Bactrocera citrus, and achieve polymorphism High, clear amplified fragments, optimized reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 extracts the DNA of Bactrocera citrus
[0023] 1 Gathering materials
[0024] The collected samples (adults and larvae) from 10 different geographical populations of Bactrocera citrus in my country were soaked in absolute ethanol and stored at -20°C. See Table 1 for information on the collection points of the population and the number of populations collected.
[0025] Table 1 Collection information of samples of Bactrocera citrus for testing
[0026]
[0027]
[0028] 2 Extraction of DNA from Bactrocera citrus
[0029] Get the citrus fruit fly worm bodies of 10 different geographical populations collected above, adopt the CTAB method to extract genomic DNA, and operate according to the following steps:
[0030] (1) Take 100 mg of the body of Bactrocera citrus, add liquid nitrogen and grind it into a powder, and quickly transfer it into a 1.5 ml Eppendorf tube.
[0031] (2) Add 800 μl of CTAB extraction buffer to the centrifuge tube in step 1, mix we...
Embodiment 2
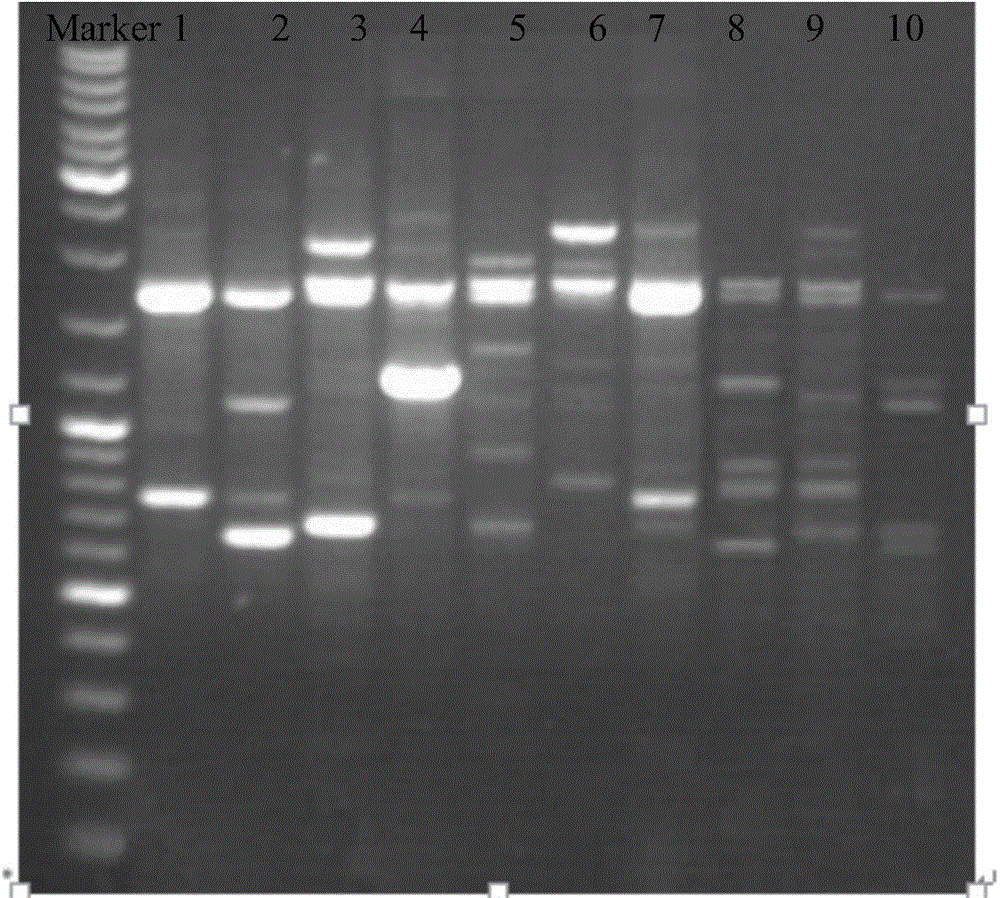
[0038] Embodiment 2RAPD analysis
[0039] Seven RAPD primers were used to perform PCR amplification on the DNA samples of 10 different geographic populations of B. citrus extracted in Example 1, and the RAPD random primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd. The names and sequences of the seven RAPD primers are as follows: 1: ACGCCAGAGG, 2: GAGCGAGGCT, 3: CAGGGGTGGA, 4: GGTCTGGTTG, S75: GACGGATCAG, Y06: AAGGCTCACC, Y18: GCTGAGTCAG. The total volume of the PCR reaction system is 25uL, including 1×PCRBuffer 2.0μL, 1.5mmol / μL MgCl 2 1.5 μL, 500 μmol / L dNTPs 1.5 μL, 0.5 μmoL / L primer 1.0 μL, 1UTaq enzyme 0.4 μL, 20ng / μL DNA template 1.0 μL, water up to 25 μL. The amplification program was: 94°C pre-denaturation for 5 minutes; 94°C for 60 s, 38°C for 60 s, 72°C for 90 s, a total of 32 cycles; 72°C for 10 min.
[0040]The amplified product was electrophoresed on 1.5% agarose gel (1×TBE buffer, 4V / cm electrophoresis), stained with ethidiu...
Embodiment 3
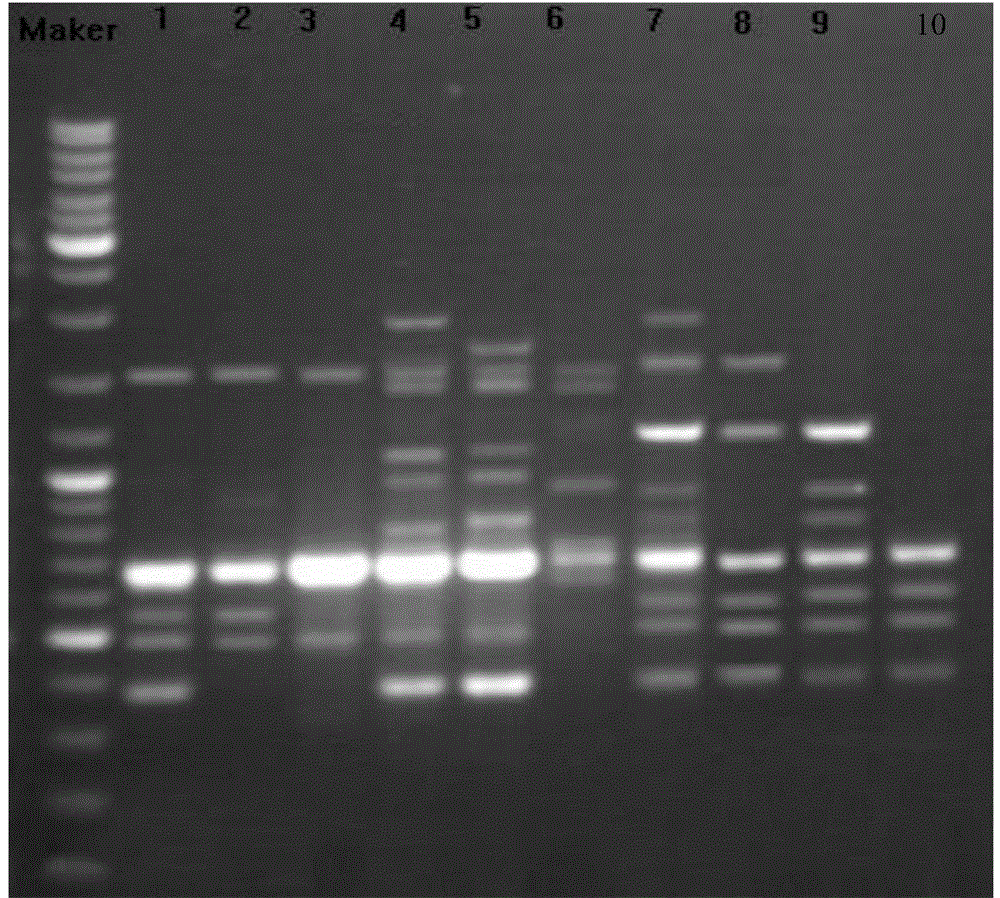
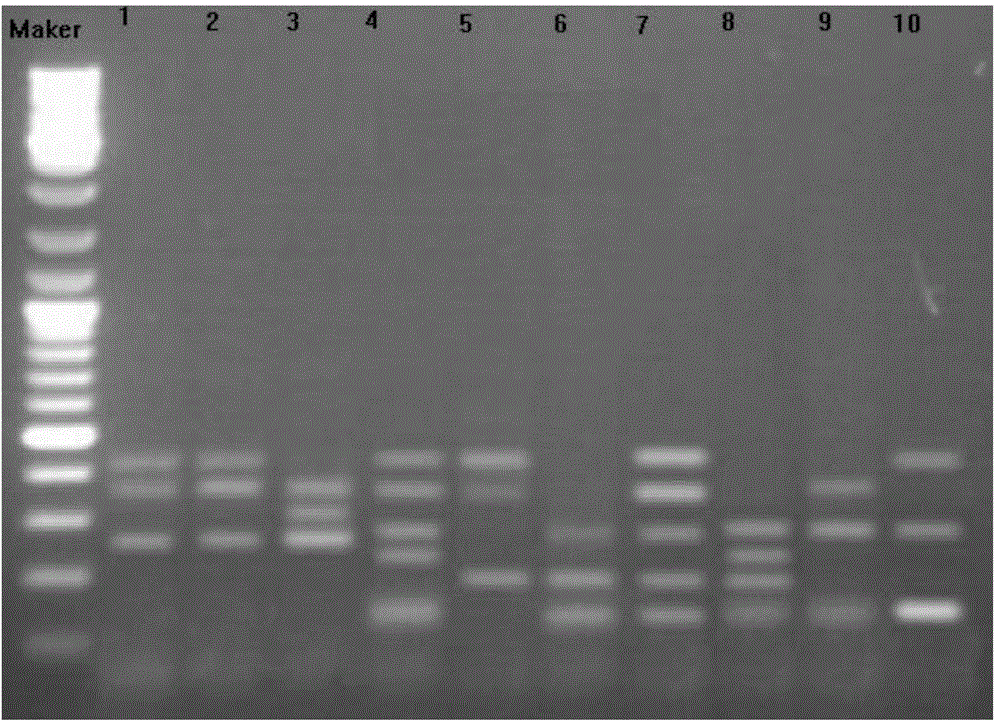
[0041] Embodiment 3SRAP analysis
[0042] 10 pairs of SRAP primer combinations were used to perform PCR amplification on the DNA samples of B. citrus of 10 different geographical populations extracted in Example 1, and the SRAP primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd. The total volume of the PCR reaction system is 25uL, including 1×PCRBuffer 2.0μL, 1.5mmol / μL MgCl 2 1.5 μL, 1.5 μL of 500 μmol / L dNTPs, 1.0 μL each of 0.5 μmol / L upstream and downstream primers, 0.4 μL of 1UTaq enzyme, 1.0 μL of 20ng / μL DNA template, and replenish water to 25 μL. Amplification program: 94°C pre-denaturation for 3 minutes; 94°C for 40s, 63°C for 40s, 72°C for 60s; cycle 15 times, each cycle annealing temperature decreased by 0.5°C; 94°C for 40s, 55.5°C for 40s, 72°C for 60s, cycle 25 times ; 72°C extension for 10 min. The 10 pairs of SRAP primer combinations are named: 3F-1R, 3F-2R, 9F-1R, 9F-3R, 9F-2R, 9F-11R, 4F-10R, 12F-11R, 10F-3R, 10F-10R ; Wh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com