Application of reagent for increasing NO content of stigmas for improving selective fertilization of cruciferous plants and improvement method
A cruciferous, selective fertilization technology, applied in the fields of botanical equipment and methods, plant genetic improvement, application, etc., can solve problems such as incompatibility of distant hybridization, and improve the selective fertilization and application scope of cruciferous plants. Wide and less harmful effect on plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
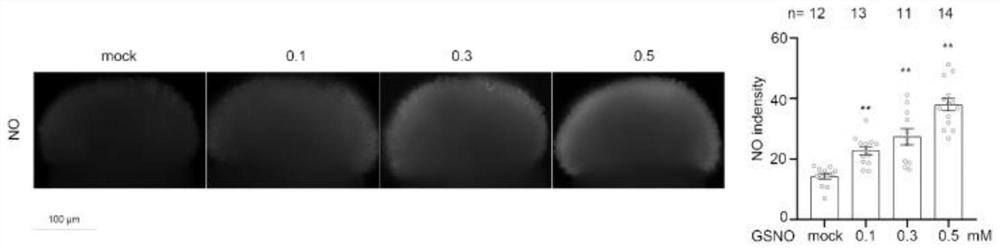
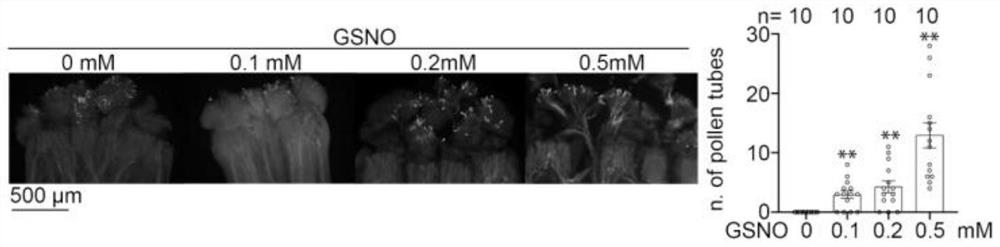
[0047] Chinese cabbage removes the flowers that have opened one day before adding GSNO, and collects the seed bags. The next morning, add GSNO solution twice to the newly opened flowers (the GSNO solution contains 0.0125% Tween20, and the GSNO concentration is 0mM, 0.1mM, 0.2mM, 0.5mM), with an interval of 30min, and the second drop After self-pollination, the nesting belts are used to prevent contamination of other pollen. After 12 days, the fruit pods were dissected, and the number of enlarged ovules was counted.
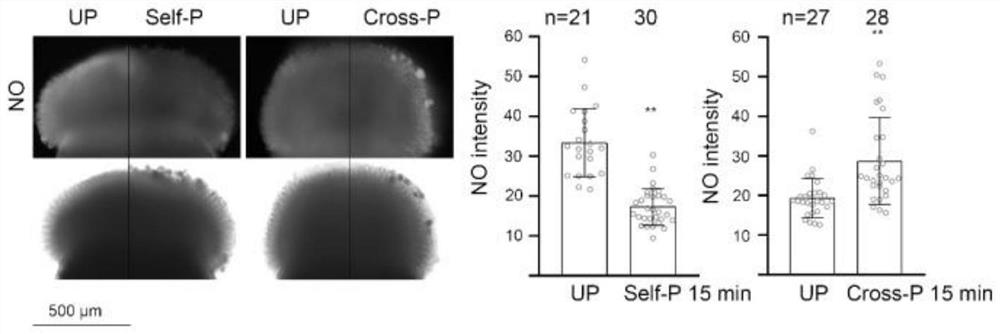
[0048] The result is as Figure 2 to Figure 5 As shown in Table 1, submission of GSNO significantly increased the stigma NO content of Chinese cabbage (for details, see figure 2 ), so that the average number of pollen tubes passing through each cabbage stigma increased from 0 to 2.93, 4.29, 12.93 (see image 3 ); Make the fruit pod length of 12 days after selfing of Chinese cabbage improve respectively from original 9.11mm to 25.20mm, 32.80mm, 32.80mm (for det...
Embodiment 2
[0052] Chinese cabbage removes the flowers that have opened one day before adding GSNO, and collects the seed bags. The next morning, emasculate the newly opened flowers and submit 2 times of GSNO solution (GSNO solution contains 0.0125% Tween20, GSNO concentration is 0mM, 0.1mM, 0.5mM, 1mM), each interval is 30min, and the second drop After the cabbage pollen is given, the seed belts are interlocked to prevent contamination of other pollen. After 12 days, the fruit pods were dissected, and the ovules in the fruit pods were counted.
[0053] The result is as Figure 6 ~ Figure 8 As shown in Table 2, after dropping GSNO, the average number of cabbage pollen tubes passing through each cabbage stigma increased from 0 to 13.75, 91.81 (for details, see Image 6 ); Make the fruit pod length of 12 days after the distant hybridization of Chinese cabbage and cabbage improve from original 0mm to 4.45mm, 24.34mm respectively (for details see Figure 7 ); after the distant hybridizatio...
Embodiment 3
[0057] Chinese cabbage removes the flowers that have opened one day before adding GSNO, and collects the seed bags. The next morning, emasculate the unopened flower buds, add 2 times of GSNO solution dropwise (GSNO solution contains 0.0125% Tween20, GSNO concentration is 0mM, 0.1mM, 0.5mM, 1mM), each interval is 30min, the second time After dripping, the pollen of the European mountain mustard was given, and the seed belt was collected to prevent other pollen from being polluted. After 12 days, the fruit pods were dissected, and the ovule situation in the fruit pods was counted.
[0058] The result is as Figure 9 ~ Figure 11 As shown in Table 3, after adding GSNO dropwise, the number of pollen tubes passing through each cabbage stigma increased from 0 to 0.50, 22.80, 58.70 (see Figure 9 ); Make the fruit pod length of 12 days after the distant hybridization of Chinese cabbage and European mountain thaliana increase to 0.95mm, 2.61mm, 12.72mm respectively from original 0mm ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com