Improved RNA editing method
An editing and targeting technology, applied in the fields of RNA editing and gene editing, can solve the problems of reduced editing efficiency and inability to edit
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
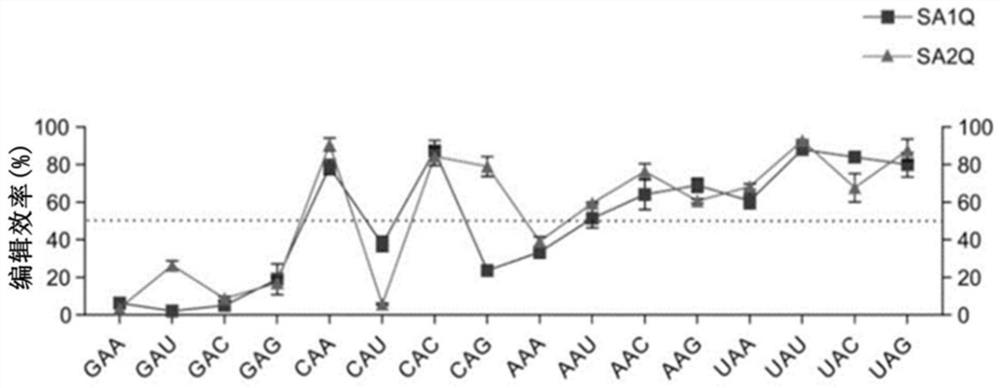
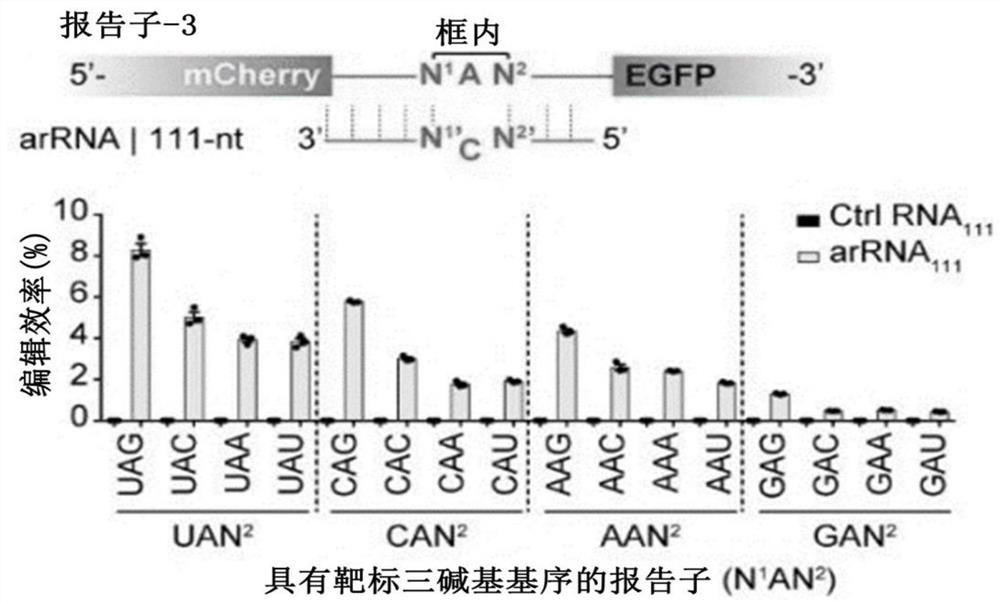
Image
Examples
Embodiment 1
[0163] Example 1: Construction of a three-base motif reporter system and corresponding arRNA
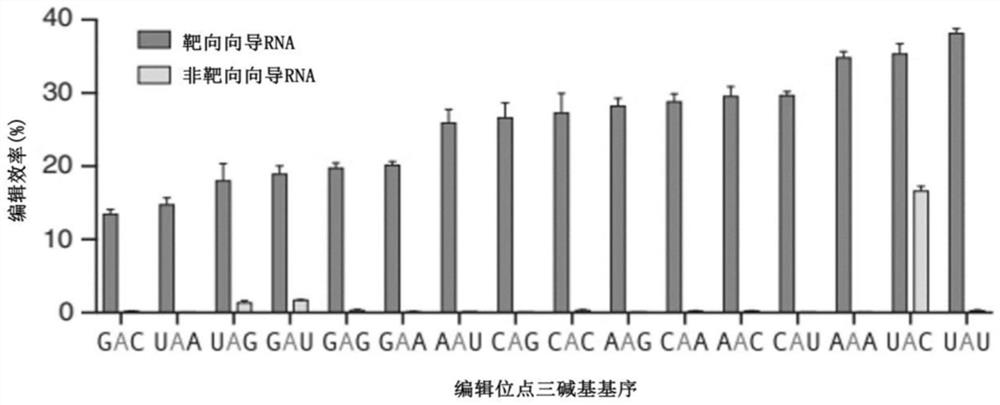
[0164] First, we constructed a reporter system containing 16 three-base motifs. Since in the LEAPER literature, the difference in editing efficiency when the three-base motif is UAG has been tested (Qu et al., 2019), in this example, in order to maintain the consistency of the control, arRNA can complement other than the editing site The paired parts all use the same sequence design as in the LEAPER literature, such as Figure 5 shown. The original plasmid Reporter1 was donated by Professor Wei Wensheng, School of Life Sciences, Peking University. The plasmid map is as follows: Figure 14 As shown, the plasmid contains the sequences shown in Table 4. Synthesize 16 kinds of three-base motif-related primers as shown in Table 1, and according to the methods well known to researchers in the field in "J. Sambrook, M.R. Green, Molecular Cloning Experiment Guide (Fourth Edition), 2017" ...
Embodiment 2
[0166] Example 2: Comparing the editing efficiency of different arRNAs to the UAG three-base motif through the positive ratio of GFP
[0167] Such as Figure 5 Shown are 16 kinds of target RNAs described in Example 1, which all have the GFP green fluorescent protein nucleic acid sequence at the 3' end of the target sequence. This sequence normally translates correctly and fluoresces green. But when the three-base motif is UAG, because UAG is a stop codon, translation will stop there, and then it cannot be translated into GFP. In this example, the A in the UAG three-base motif was edited by the LEAPER system. If the editing is successful, UAG will be converted into UIG, and UIG will be recognized as UGG during the translation process, so that the translation will not be terminated, so that the downstream GFP can be translated normally. Therefore, we can roughly judge the level of editing efficiency of different arRNAs through the size of the positive ratio of GFP.
[0168] ...
Embodiment 3
[0177] Embodiment 3: The RNA editing efficiency determination of GAN tribasic motif
[0178] In this example, 16 kinds of arRNA were transfected to the reporter system cells respectively containing the three base motifs of UAG, GAA, GAU, GAC, and GAG, and the transfection steps were the same as in Example 2.
[0179] After 72h (48h after transfection), the sample was collected by TRIZOL and RNA was extracted (TRIzol Reagent, ambientREF15596026), and 1 μg of RNA was reverse-transcribed, and the reverse transcription system was 20 μL ( One-Step gDNARemoval and cDNA Synthesis SuperMix, full gold AT311-02), take 1 μL of the reverse transcription product and perform PCR with the following pair of primers: ggagtgagtacggtgtgcGACGAGCTGTACAAGCTGCAGGG (SEQ ID NO: 1), gagttggatgctggatggTGGTGCAGATGAACTTCAGGGTCAG (lowercase The letters indicate the primer adapters required by the Hi-Tom kit) for PCR amplification and library construction with the Hi-Tom kit (Novogene, REF PT045).
[0180...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com