Method and device for integrated detection of CNV, uniparental disomy, triploidy and Roh
A uniparental disomy and triploid technology, applied in the field of genetic testing, can solve the problems of high testing cost and complicated process, and achieve the effect of simplifying the testing process, reducing the testing cost and improving the testing efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
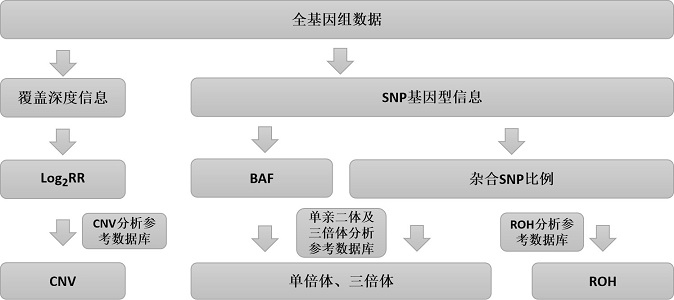
[0124] In this embodiment, the method of integrated detection of CNV, uniparental disomy, triploidy and ROH of the present invention is used to detect small CNV samples (cell line samples), including the following steps:
[0125] 1. Construction of genome sequencing library;
[0126] 2. Sequencing on the machine to obtain the whole genome sequencing data of the processed sample 1;
[0127] 3. Obtain analysis parameters: coverage depth information and SNP genotype information of each window;
[0128] 4. Based on the sequencing depth information, follow the CNV analysis process to obtain small CNVs of more than 100 kb;
[0129] 5. Based on the SNP genotype information, detect uniparental disomy and polyploidy according to the uniparental disomy / triploid analysis process;
[0130] 6. Based on the SNP genotype information, detect more than 5M ROH according to the ROH analysis process.
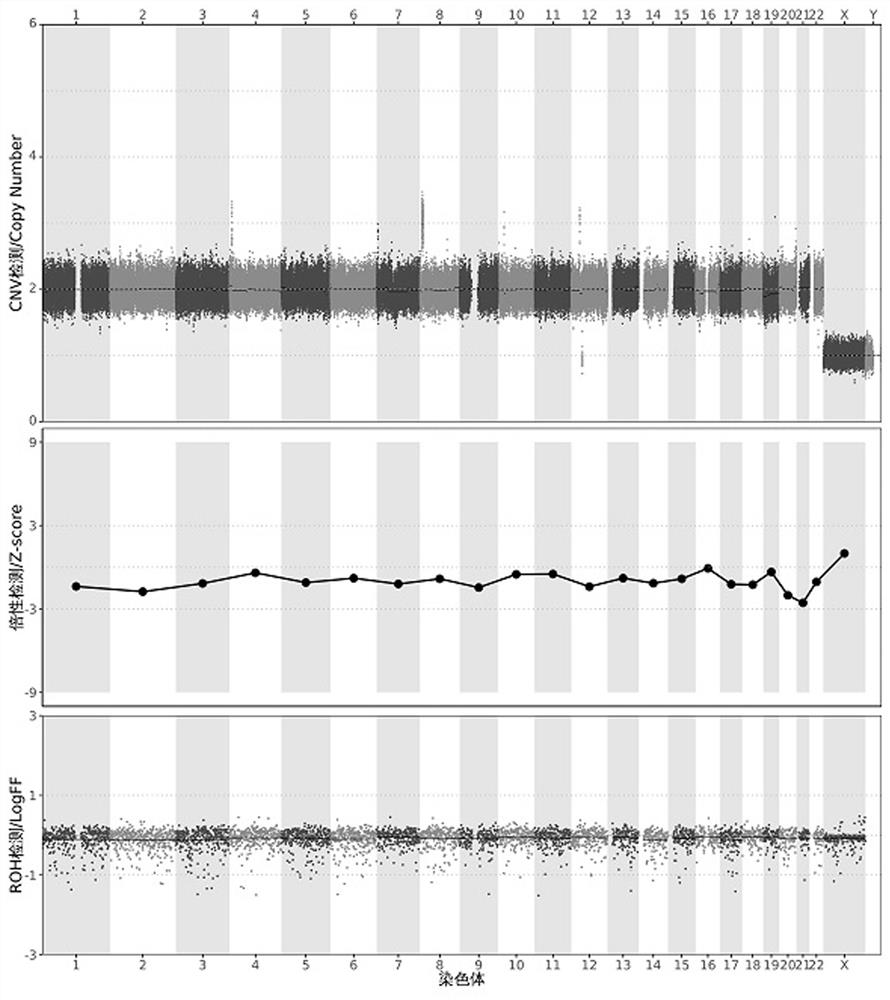
[0131] Test results such as figure 2 As shown, the CNV detection result is:
[0132] dup(...
Embodiment 2
[0138] In this embodiment, the method for the integrated detection of CNV, uniparental disomy, triploidy and ROH of the present invention is used to detect uniparental disomy samples (cell line samples), including the following steps:
[0139] 1. Construction of genome sequencing library;
[0140] 2. Sequencing on the machine to obtain the whole genome sequencing data of the processed sample 2;
[0141] 3. Obtain analysis parameters: coverage depth information and SNP genotype information of each window;
[0142] 4. Based on the sequencing depth information, follow the CNV analysis process to obtain small CNVs of more than 100 kb;
[0143] 5. Based on the SNP genotype information, detect uniparental disomy and polyploidy according to the uniparental disomy / triploid analysis process;
[0144] 6. Based on the SNP genotype information, detect more than 5 M ROH according to the ROH analysis process.
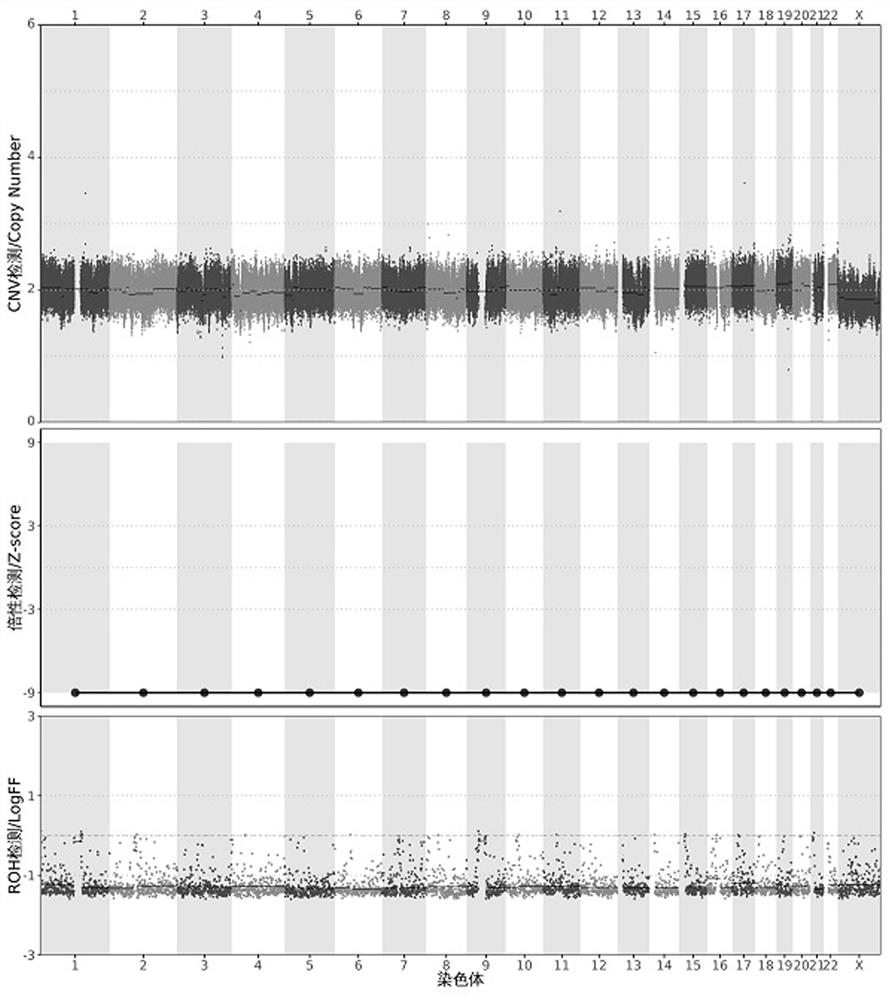
[0145] Test results such as image 3 As shown, CNV detection results: no CNV...
Embodiment 3
[0147] In this embodiment, the method of integrated detection of CNV, uniparental disomy, triploid and ROH of the present invention is used to detect triploid samples (cell line samples), including the following steps:
[0148] 1. Construction of genome sequencing library;
[0149] 2. Sequencing on the machine to obtain the whole genome sequencing data of the processed sample 3;
[0150] 3. Obtain analysis parameters: coverage depth information and SNP genotype information of each window;
[0151] 4. Based on the sequencing depth information, follow the CNV analysis process to obtain small CNVs of more than 100 kb;
[0152] 5. Based on the SNP genotype information, detect uniparental disomy and polyploidy according to the uniparental disomy / triploid analysis process;
[0153] 6. Based on the SNP genotype information, detect more than 5 M ROH according to the ROH analysis process.
[0154] Test results such as Figure 4As shown, CNV detection results: +(mosaic)(X); -(mosaic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com