Tataxin-sensitive markers for determining efficacy of ataxin replacement therapy
A marker and therapy technology, applied in the direction of determination/testing of microorganisms, microorganisms, measuring devices, etc., can solve the problem of antioxidants and iron chelation not being very effective
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0358] Example 1: Generation of FXN-induced signatures
[0359] FXN fusion protein
[0360] The FXN fusion protein used in this example is a fusion protein comprising TAT-cpp and hFXN linked by a linker at the N-terminus of hFXN (Vyas et al. (2012) Hum. Mol. Genet. 21, 1230-1247) , referred to herein as CTI-1601. The hFXN in the fusion protein is the full-length 210 aa long precursor form of cotaxin, which contains an 80 aa mitochondrial targeting sequence (MTS) at the N-terminus. The full-length hFXN protein (amino acids 1-210) includes the amino acid sequence of SEQ ID NO:1.
[0361]
[0362]
[0363] When the protein is imported into the mitochondrial matrix, it is cleaved at amino acid 81 to generate the mature form of FXN, resulting in mature active FXN of 130 amino acids with a predicted molecular weight of 14.2 kDa (SEQ ID NO: 1).
[0364]
[0365] Full length hFXN (SEQ ID NO: 1 ) includes mature hFXN (SEQ ID NO: 2) and a mitochondrial targeting sequence (...
Embodiment 2
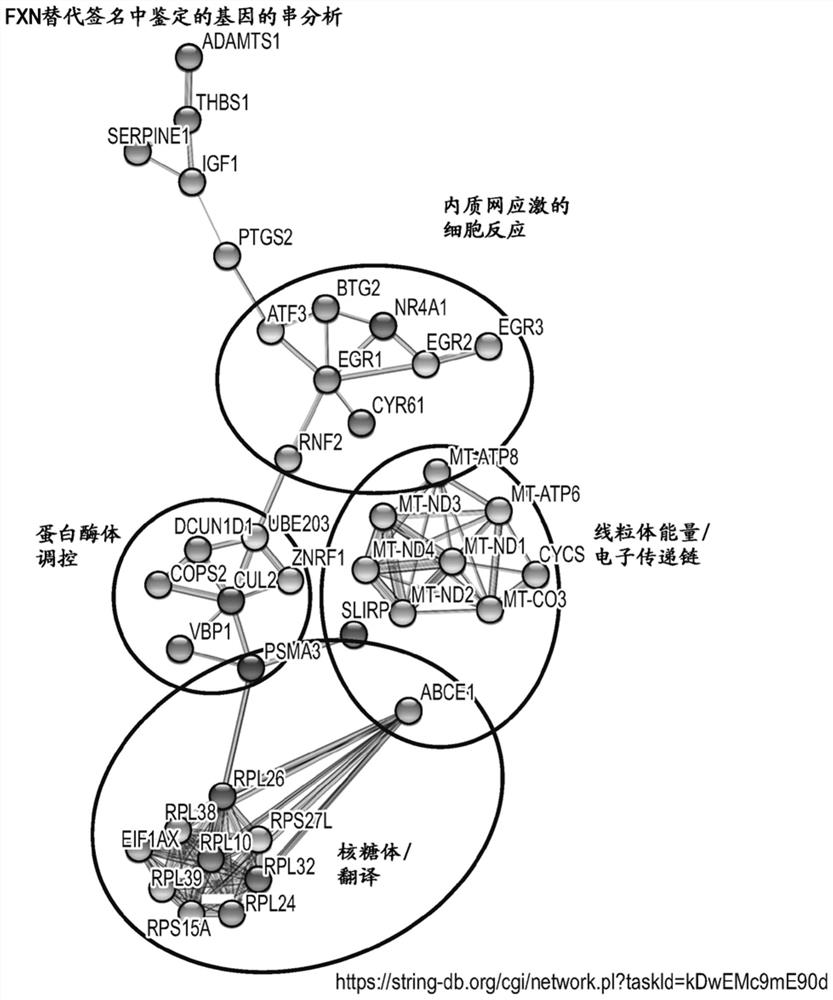
[0409] Example 2: String analysis of FSGM
[0410] This example describes string analysis of the FSGMs shown in Table 2, showing that the protein products of the FSGMs are at least partially biologically connected as a group.
[0411] String analysis was performed using the 85 protein products of the FXN-sensitive genomic markers described in Table 2 using the string database (string-db.org; Szklarczyk et al. (2015) DOI: 10.1093 / nar / gkv1277 and references therein) . string analysis such as figure 1 shown. String analysis represents examples of known and / or predicted protein interactions based on their function. The parameters used to generate clusters in string analysis were: nodes = 85; edges = 97; average node degree = 2.28; average local clustering coefficient = 0.345; expected number of edges = 35; PPI enrichment p-value figure 1 Only some clusters are clearly visible in , while others are not visible in the figure, but these are also listed below in this paper.
[0...
Embodiment 3
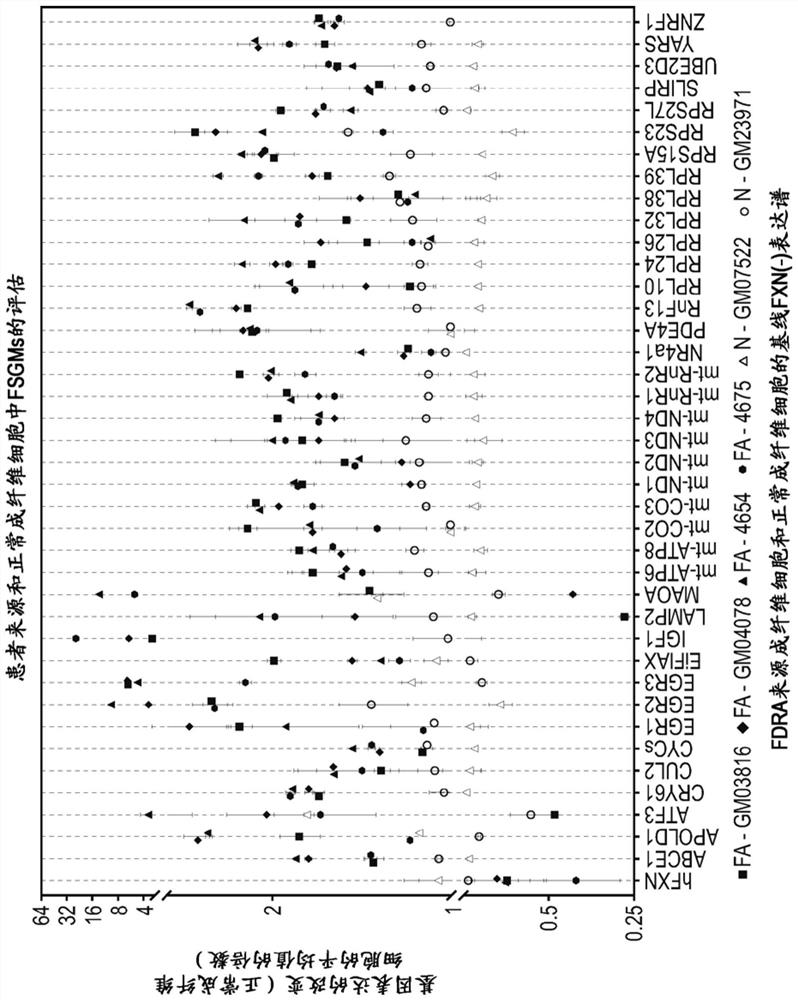
[0427] Example 3: Selection of potential FXN target genes after in vitro treatment
[0428] The identification of genes that are inversely regulated by FXN gene ablation followed by FXN protein replacement in vivo suggests that changes in gene expression induced by FXN replacement therapy can be used as an indicator of therapeutic efficacy in patients treated with FXN replacement therapy. Based on this premise, the baseline FXN-induced signature was tested in two in vitro human cell models: Friedreich's ataxia (FDRA)-derived fibroblasts and in
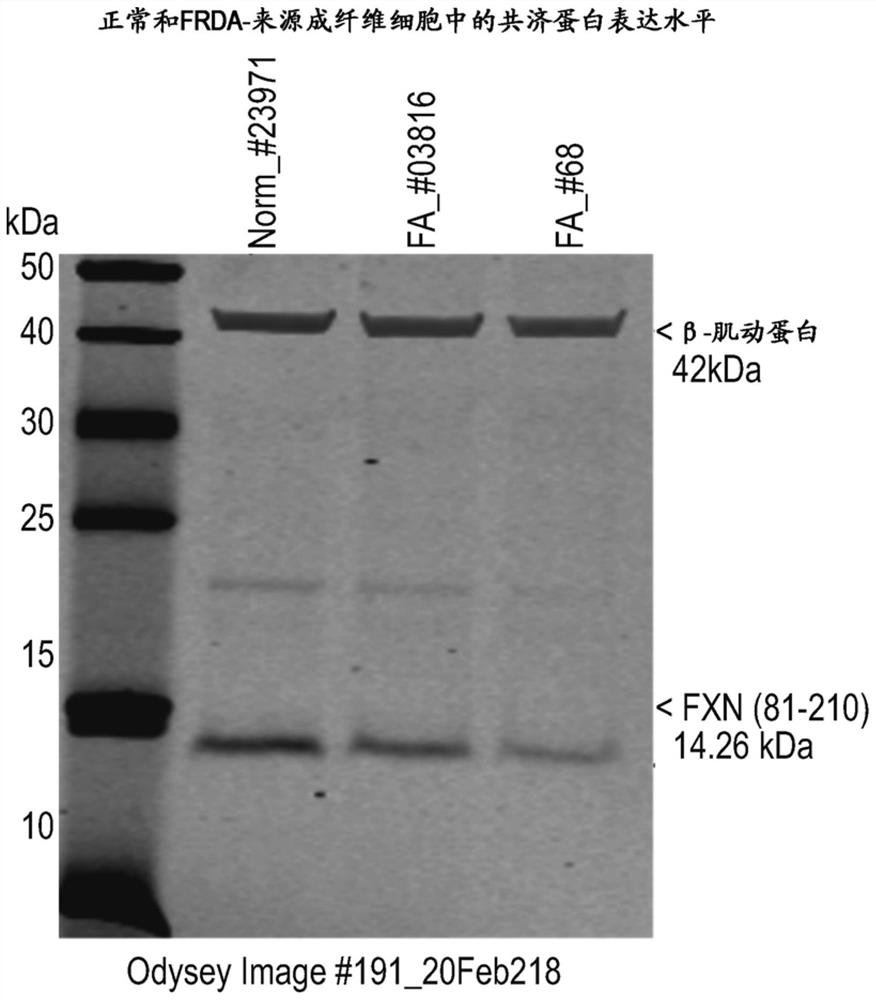
[0429] Musclein protein and mRNA expression in human cell models
[0430] Detection of frataxin in FDRA-derived fibroblasts proteinand mRNA expression. Western blot gels visualized and quantified frataxin protein expression, while frataxin mRNA expression was quantified by qRT-PCR. The results are shown in figure 2 and Table 3.
[0431] figure 2 Fraxin detection in control GM23971 cells and FDRA-derived fibroblasts FAGM03816...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com