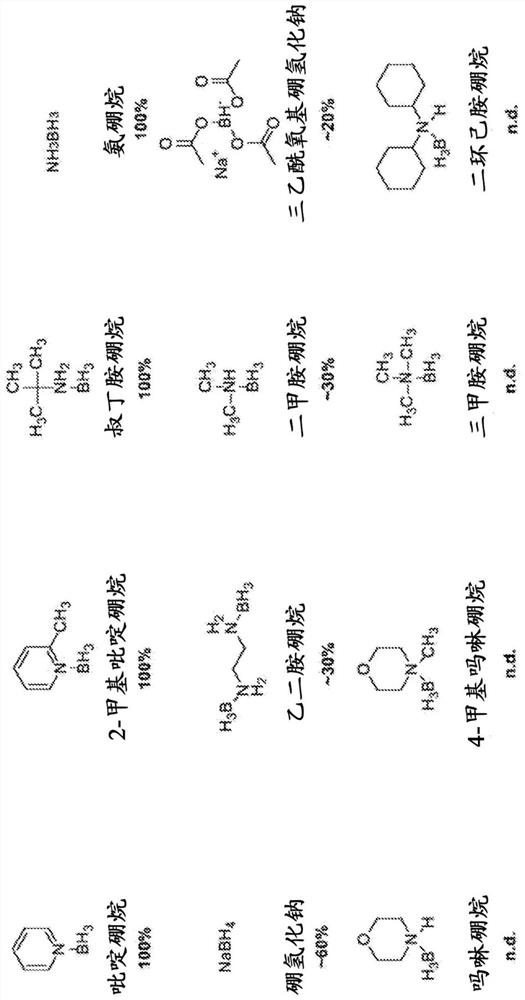
Hydrosulfite-free whole genome methylation analysis
A technology of hydroxymethylcytosine and methylcytosine, which is applied in the field of bisulfite-free genome-wide methylation analysis, can solve the problem that methylation pictures cannot be generated
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0247] Example 1: TAPS and WGTAPS
[0248] method
[0249] Preparation of model DNA.
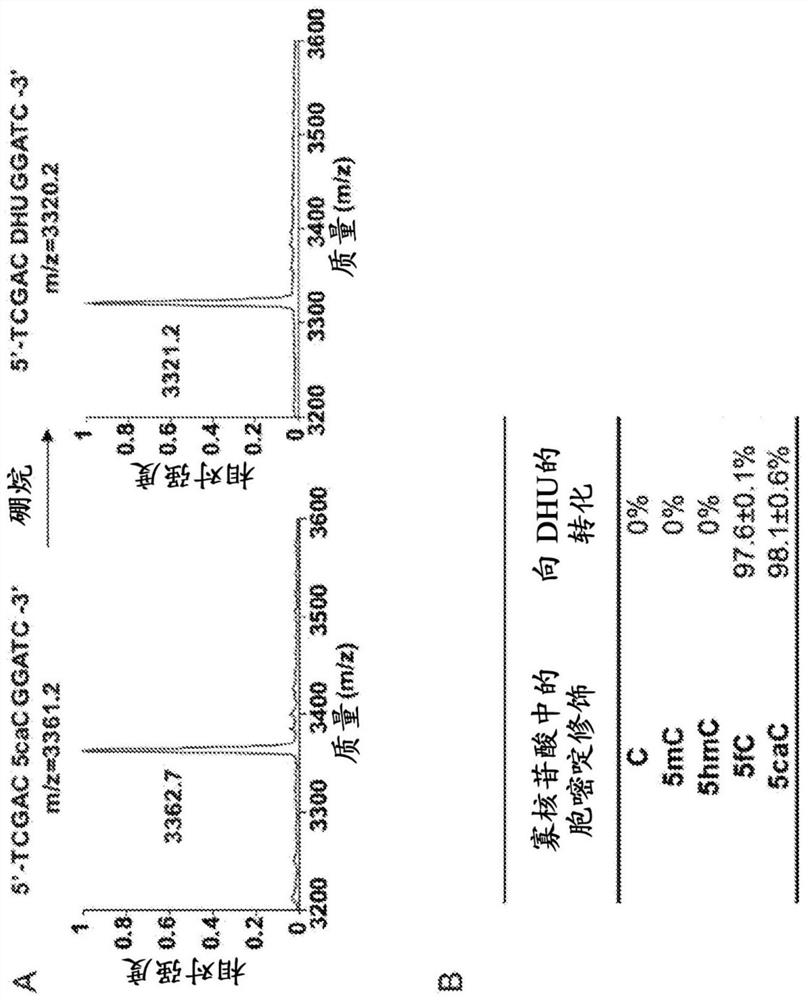
[0250] DNA oligonucleotides for MALDI and HPLC-MS / MS testing. DNA oligonucleotides ("oligonucleotides") with C, 5mC and 5hmC were purchased from Integrated DNA Technologies (IDT). All sequences and modifications can be found at Figure 6 and Figure 7 middle. DNA oligonucleotides with 5fC were synthesized by the C-tailing method: DNA oligonucleotides 5′-GTCGACCGGATC-3′ were annealed to 5′-TTGGATCCGGTCGACTT-3′ followed by 5-formyl-2′-dCTP (Trilink Biotech) and Klenow fragment 3'→5'exo-(New England Biolabs) were incubated in NEB buffer 2 at 37°C for 2 hours. The product was purified with a Bio-Spin P-6 gel column (Bio-Rad).
[0251] DNA oligonucleotides with 5caC were synthesized using the Expedite 8900 DNA synthesis system using the standard phosphoramidite (Sigma) 5-carboxy-dC-CE phosphoramidite (Glen Research). Subsequent deprotection and purification were performed using Glen-Pak...
Embodiment 2
[0385] Example 2: Endonuclease Enriched TAPS (eeTAPS)
[0386] method
[0387] Preparation of Spike Controls.
[0388] 4 kb were prepared by PCR amplification of the pNIC28-Bsa4 plasmid (Addgene, Cat #26103) in reactions containing 1 ng DNA template, 0.5 μM primers, and 1X Phusion High-Fidelity PCR Master Mix (Thermo Scientific) with HF buffer Spike controls. Primer sequences are listed in Table 10. PCR products were purified by Zymo-IC columns (Zymo Research) and methylated with HpaII methyltransferase (New England Biolabs) in 50 μL reactions at 37° C. for 2 hours. Methylated products were purified with 1X Ampure XP beads (Beckman Coulter) according to the manufacturer's protocol. By using M . SssI enzyme (New England Biolabs) methylated unmethylated λ-DNA (Promega) to prepare fully CpG methylated λ-DNA.
[0389] Preparation of vector DNA.
[0390] Vectors were prepared by PCR amplification of the pNIC28-Bsa4 plasmid (Addgene, catalog #26103) in reactions containing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com