SNP site combination for colorectal cancer onset risk prediction, onset risk prediction model and system
A colorectal cancer and risk prediction technology, applied in the field of biomedicine, can solve the problems of unclear screening effect and no genetic prediction model for the incidence of colorectal cancer in the general population.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1 SNP site combination and model construction related to colorectal cancer risk prediction model
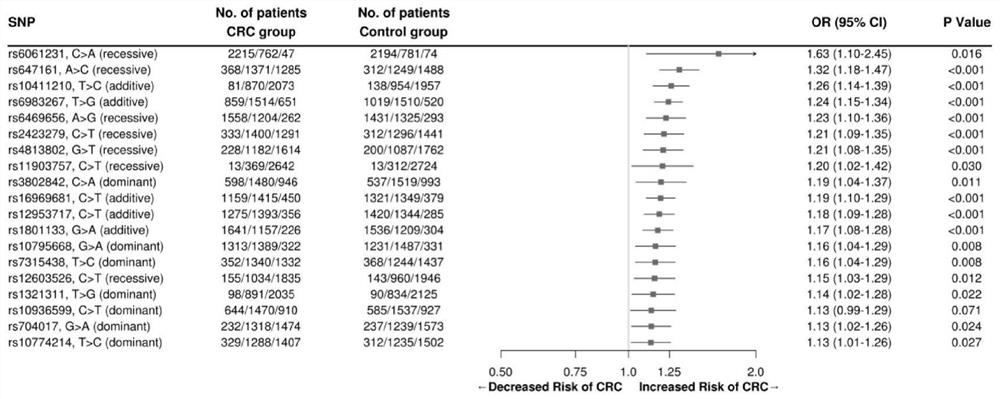
[0052] Based on the 75 key susceptibility gene mutation sites found in international colorectal cancer GWAs studies (Table 1), the large-scale sample size of Chinese colorectal cancer population and healthy control population (3049 cases of colorectal cancer population cohort in southern China) Cancer patients and 2557 healthy control individuals) were based on the association analysis, using the LASSO statistical method to perform fitting analysis on 75 SNPs, using 10-fold cross-validation to train the model, and randomly selecting the test data set each time.
[0053] Table 1. The prediction model invented by the present invention is based on 75 SNPs derived from GWAs
[0054]
[0055]
[0056]
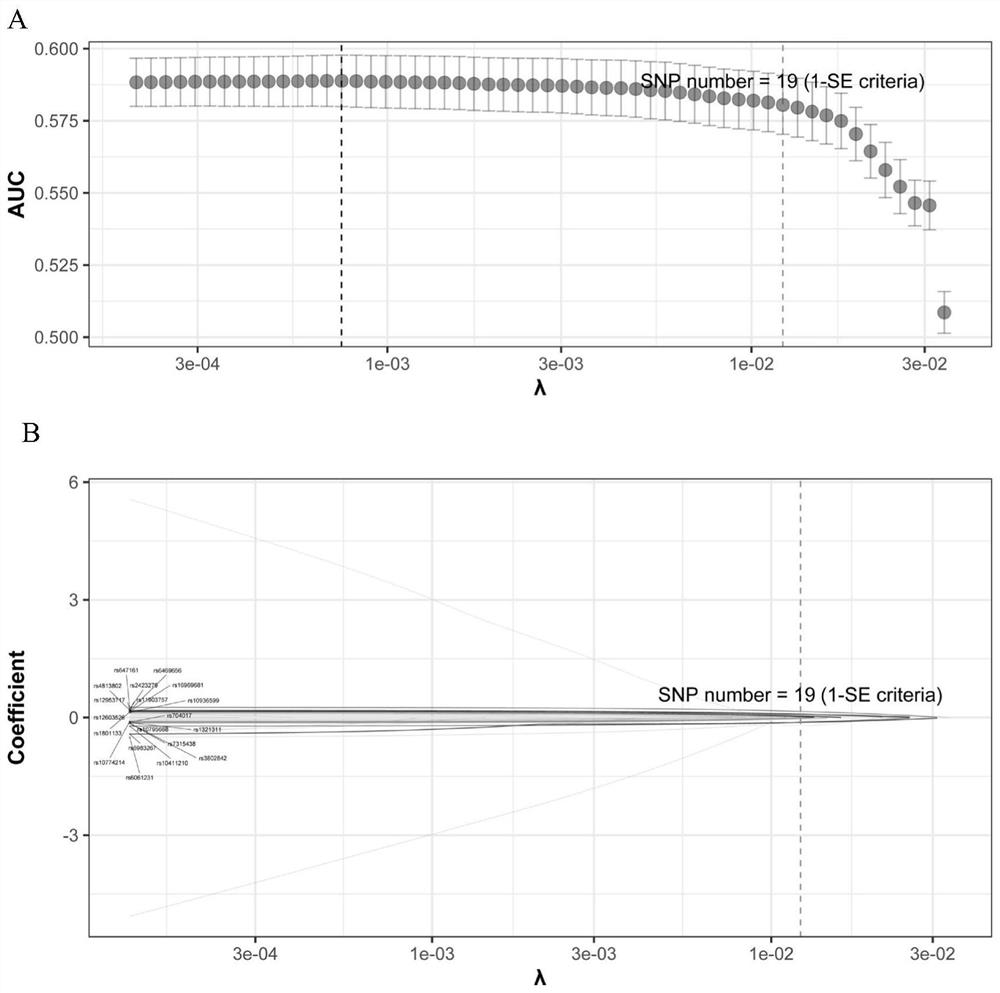
[0057] The present invention fits models containing different numbers of SNPs in the analysis process, and finally obtains an optimized colorectal cancer risk p...
Embodiment 2
[0064] Example 2 Colorectal Cancer Risk Prediction Kit
[0065] A kit for predicting the risk of colorectal cancer, comprising: PCR amplification primers and single-base extension primers for detecting 19 SNP sites in the human genome (the 19 SNP sites shown in Table 2).
[0066] The preparation of the specific kit is as follows:
[0067] 1. Design and synthesize PCR-specific recognition primers and extension primers of the SNP site, as shown in Table 3
[0068] Table 3 PCR amplification primers (Primer Allele_FAM, Primer Allele_HEX) and co-extension primers (Primer_Common) for specific recognition of biallelic genes at 19 SNP sites to be tested
[0069]
[0070]
[0071]
[0072] 2. Construction of the kit
[0073] Other components of the kit, including: Taq enzyme, dNTP mixture, diluent, buffer, etc., see the detection method below for details.
[0074] 3. The detection method, the process is as follows:
[0075] (1) DNA sample extraction
[0076] Genomic DNA w...
Embodiment 3
[0088] Embodiment 3 The internal verification of the forecasting performance of the forecasting model constructed by the present invention
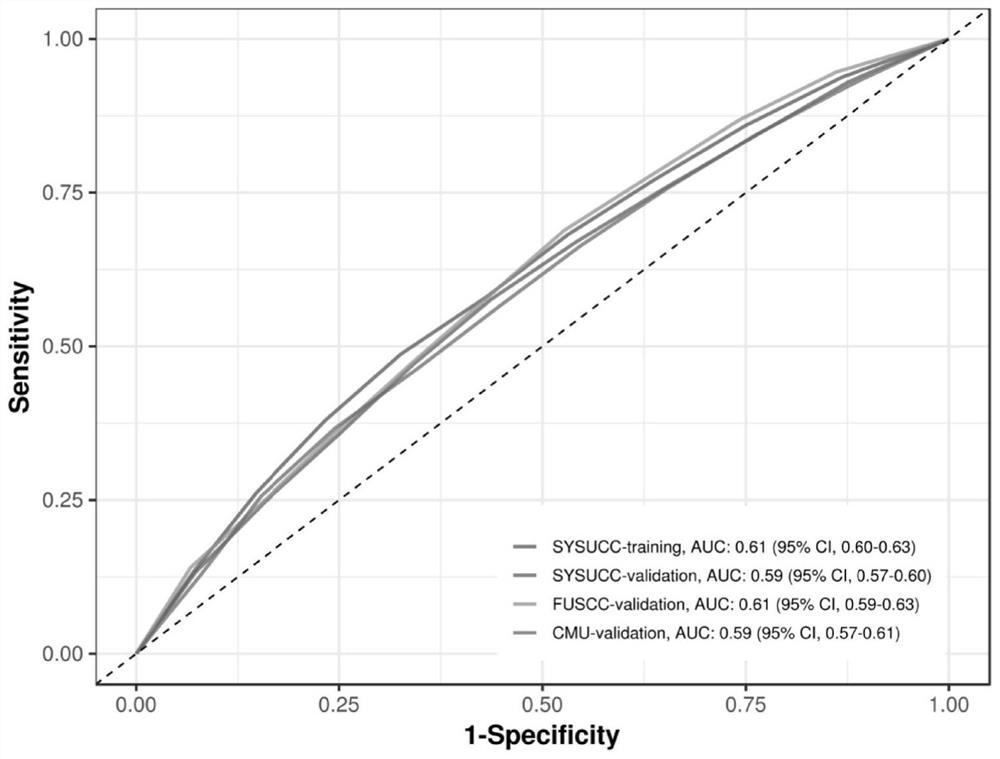
[0089] The prediction performance of the model constructed by the present invention has been confirmed in an independent verification cohort from southern China. The verification cohort is 3017 cases of colorectal cancer patients and 2488 cases of healthy control individuals. The AUC of the prediction risk of colorectal cancer is 0.59 ( image 3 ), showing that the model has good internal validity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com