Colorectal adenoma biomarker, kit and screening method of biomarker
A technology of biomarkers and screening methods, applied in the screening of kits and biomarkers, in the field of colorectal adenoma biomarkers, to achieve high specificity, high accuracy, and good screening efficiency and accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Screening of markers for early screening of colorectal adenoma based on microbial SNVs and construction of early screening models
[0058] 1.1. Data collection
[0059] From the National Center for Biotechnology Information SRA database (URL: https: / / www.ncbi.nlm.nih.gov / sra) and the European Bioinformatics Institute ENA database (URL: https: / / www.ebi.ac.uk / ena) Obtain the fecal microbial metagenomic sequencing data and clinical information data of colorectal adenoma patients and healthy control samples (clinical information mainly includes: disease status, age, gender and BMI).
[0060] The cohorts included in this embodiment are: Japanese JAP cohort (DRA006684, DRA008156), Austrian AUS cohort (ERP008729), French FRA cohort (ERP005534) and Italian ITA cohort (SRP136711), and the number of samples included in the actual analysis is 622, including 183 colorectal adenoma samples and 439 healthy control samples.
[0061] 1.2. Data preprocessing
[0062] Quali...
Embodiment 2
[0086] Example 2 Different cohort cross-validation and leave-one-out method validation
[0087] Experimental materials: Cross-validation and leave-one-out validation were performed using cohorts from different countries in public data to test the robustness and versatility of microbial biomarkers.
[0088]experimental method:
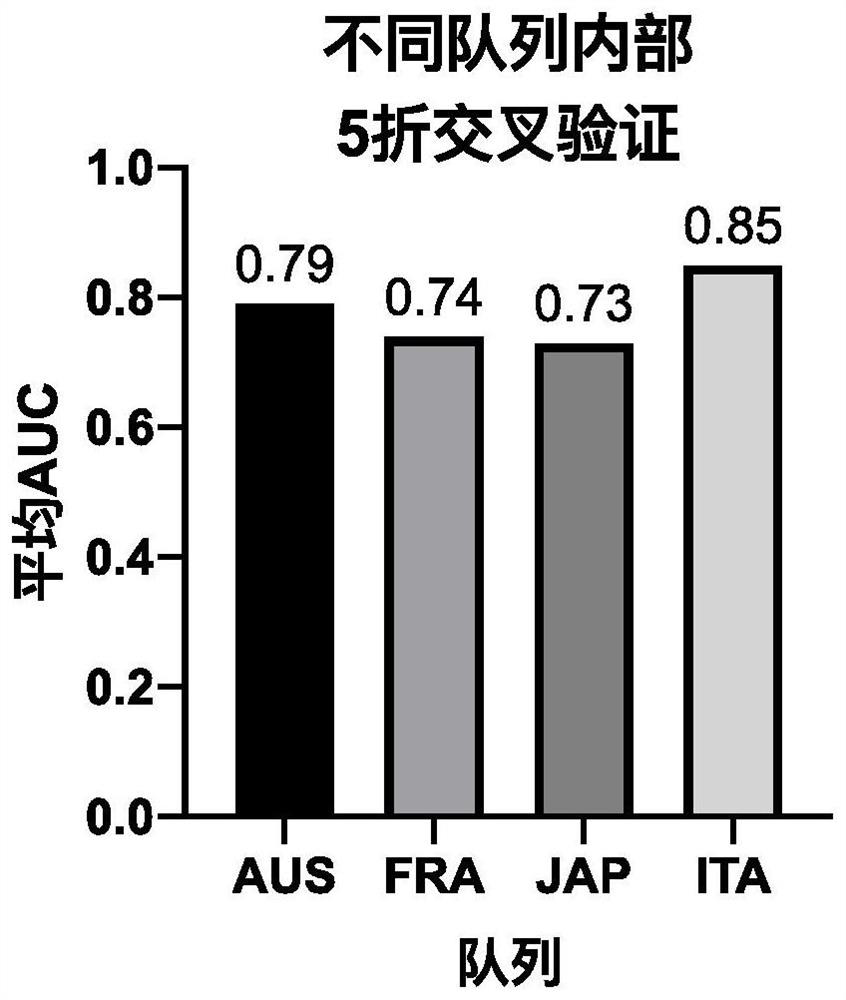
[0089] 2.1. 5-fold cross-validation within different cohorts
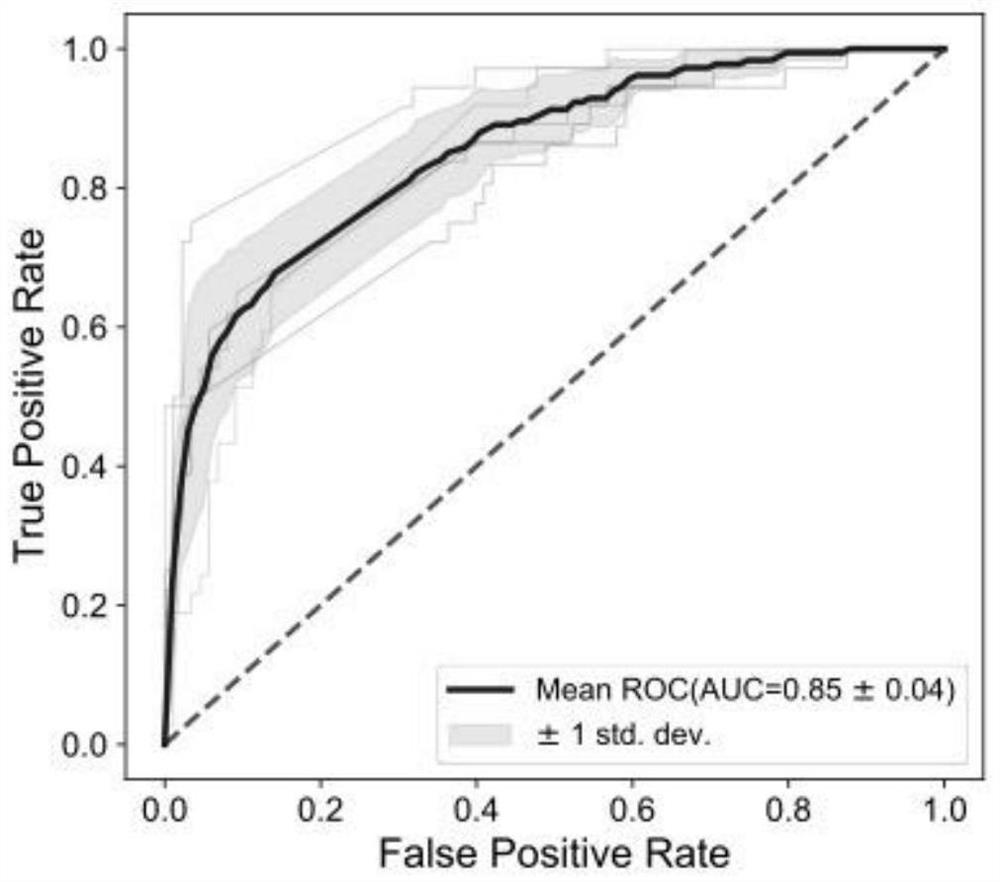
[0090] For cohorts from different countries, based on our identified optimal strain SNV combinations (E.coli_58110, F.prausnitzii_57453, F.prausnitzii_61481, P.distasonis_56985, F.prausnitzii_62201, B.longum_57796, B.adolescentis_56815, A.hadrus_55206 .uniformis_57318, B.vulgatus_57955, 10 strains), and internal 5-fold cross-validation for each queue, that is, each queue is randomly divided into 5 folds, and each fold is used as a test set in turn, and the remaining 4 folds are used as a training set Perform model building to obtain an average AUC with a 50% discount.
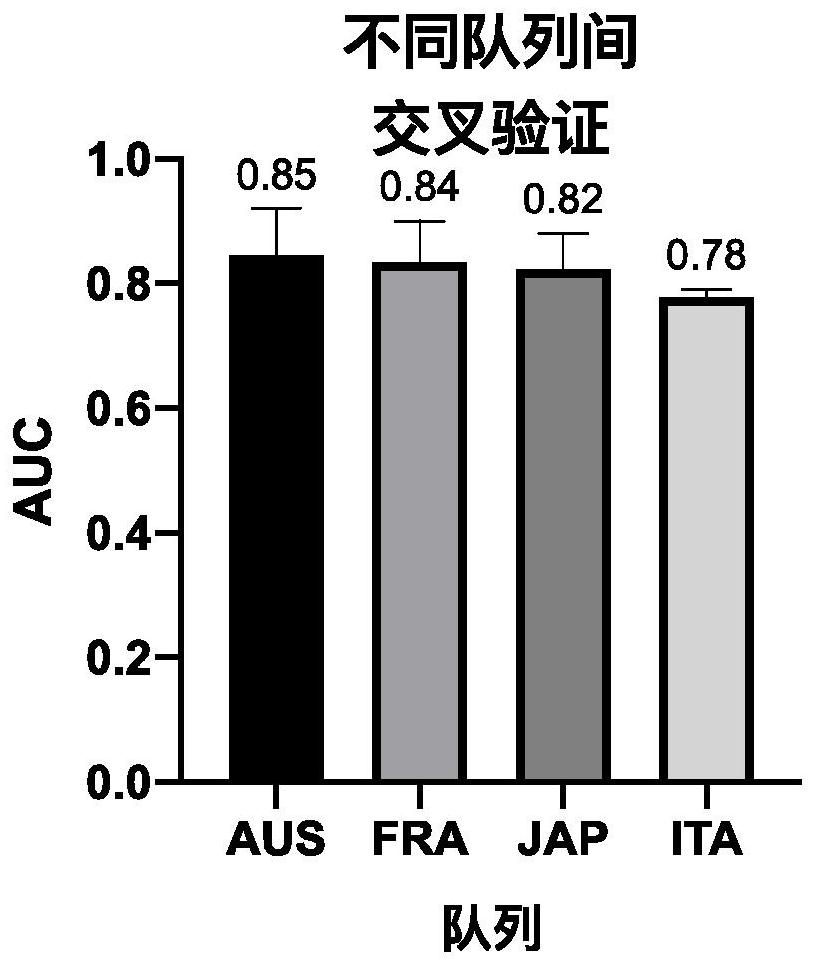
[0091] 2.2. Cross-...
Embodiment 3
[0096] Example 3 specificity verification
[0097] Experimental materials: Collect the microbiological sequencing data of intestinal diseases other than colorectal adenoma in the database for specific verification, including colorectal cancer (cohort ERP008729, ERP005534, DRA006684, DRA008156, SRP136711, the number of disease samples is 386, healthy controls The number of samples is 439), Crohn's disease (cohort PRJNA400072, the number of disease samples is 68, the number of healthy control samples is 34) and ulcerative colitis (cohort PRJNA400072, the number of disease samples is 53, the number of healthy control samples is 34).
[0098] Experimental method: For the sequencing data of different diseases, based on the optimal strain SNV combination we confirmed, we constructed a model for each disease, and obtained the result of 10-fold cross-validation, that is, each disease data was randomly divided into 10 folds internally. One fold is used as a test set in turn, and the re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com