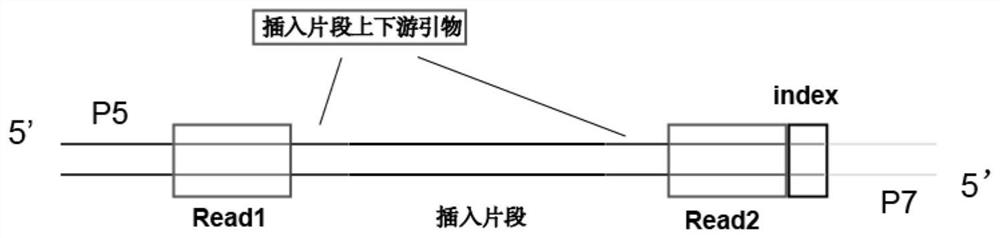
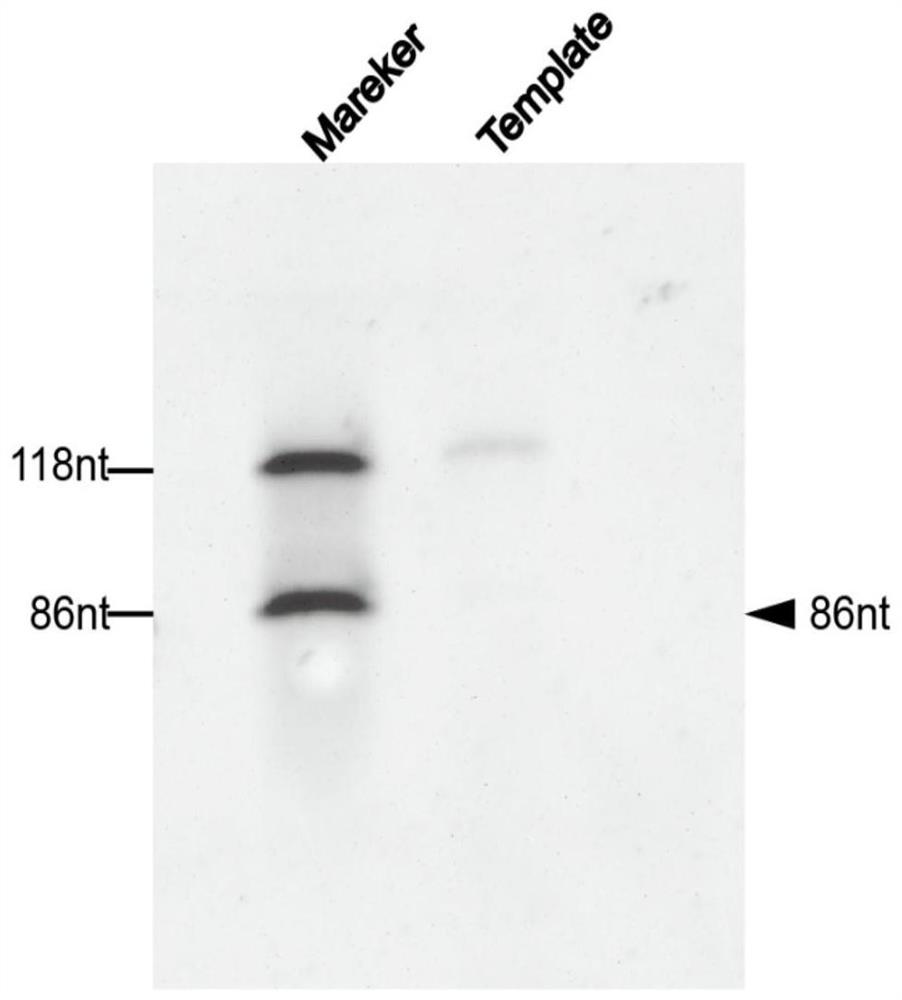
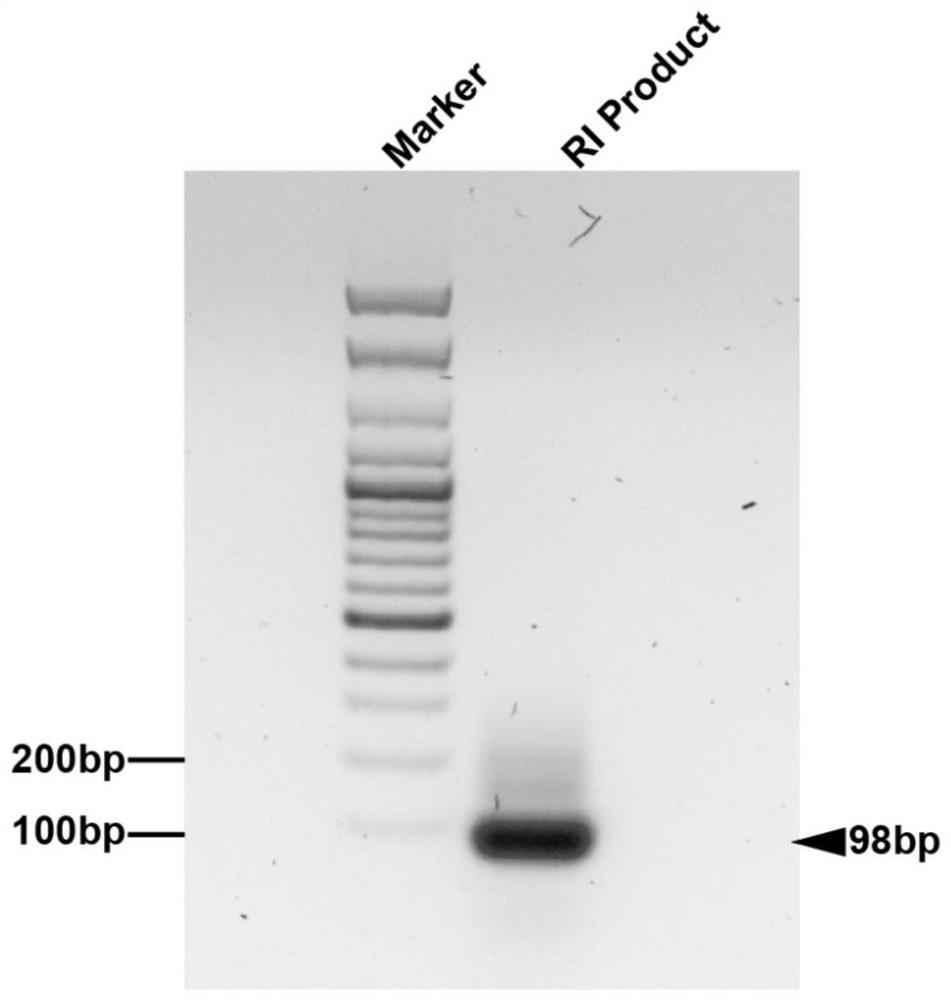
Aptamer screening method
An aptamer and random sequence technology, applied in biochemical equipment and methods, microbial measurement/testing, DNA preparation, etc., can solve the problems of time-consuming, expensive, cumbersome SELEX process, etc., and achieve the effect of simplifying experimental design
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Insert a random sequence into the non-cutting functional part of the self-cleaving DNA self-cleaving enzyme, destroy its original shearing secondary structure, and make the random sequence form a specific secondary structure under the action of the target molecule, making the DNA self-cleaving. Cleavage occurs after lyase restores the cleavage structure. The random library is:
[0032] I-R3-random: 5’-GTAACGTAGTTGAGCTG-N60-TGACGTTGAAGCGTTACGCAGCTGTGGGTTGATTCC-3’
[0033] II-R1-ramdom: 5'biotin-CATGACCACTAGGAGCATCTTTGGCG-N47-TAGGGGAATAAATCTTTGGCACCTAGTGGTCATG-3'
[0034] Among them, N60 and N47 represent 60 or 47 random bases, respectively.
[0035] The specific steps include the following:
[0036] 1. Screening of R1-random random library
[0037] 1. Preparation of I-R3 DNA random library. The ordered random library was scaled to 100 μM and then bound to the 5’ end biotin-modified oligonucleotide sequence (5’ oligo) sequence. Make the library fold correctly.
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com