Sequence sequence repeats (SSR) molecular marker of thoracic window scirpus as well as primer combination and application thereof
A technology of molecular markers and primer combinations, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve problems such as the development of molecular markers without firefly SSR
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] 1. Genome-wide SSR analysis and primer design of P.
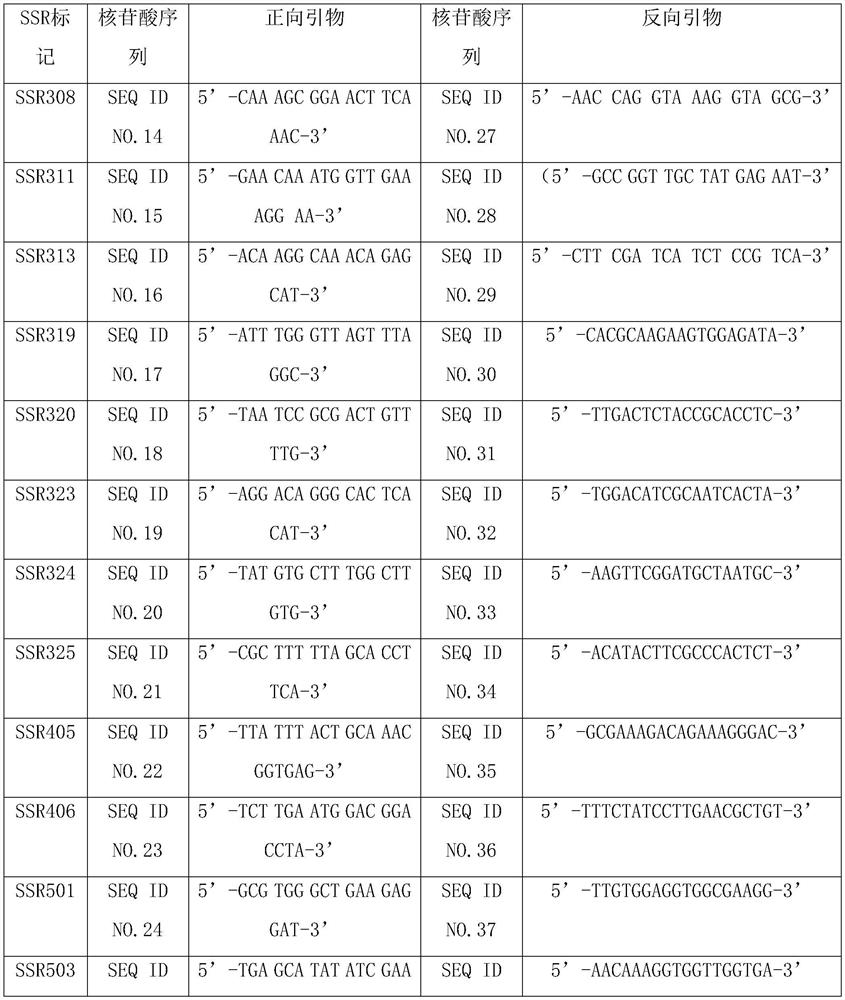
[0052] The thoracic window firefly genome was de novo sequenced, and the assembled scaffold file was used as the input file. MISA was used to perform SSR scanning of the whole genome, and primer3 was used to design primers based on the 100 bp upstream and downstream flanking sequences of the detected SSR. Primer design conditions are as follows: optimal length 24bp; shortest length 20bp; longest length 28bp; optimal annealing temperature 63°C; minimum annealing temperature 60°C; maximum annealing temperature 65°C; °C. The number and types of SSR loci were analyzed, and then high-efficiency SSR primers that could be used for the identification of different male paternity were designed and developed. Considering the conservation and stability, the applicant mainly used microsatellites with 8 to 15 repeats of three bases, four bases and five bases. A total of 44 primer pairs were designed to identify SSR
[0053] 2. ...
Embodiment 2
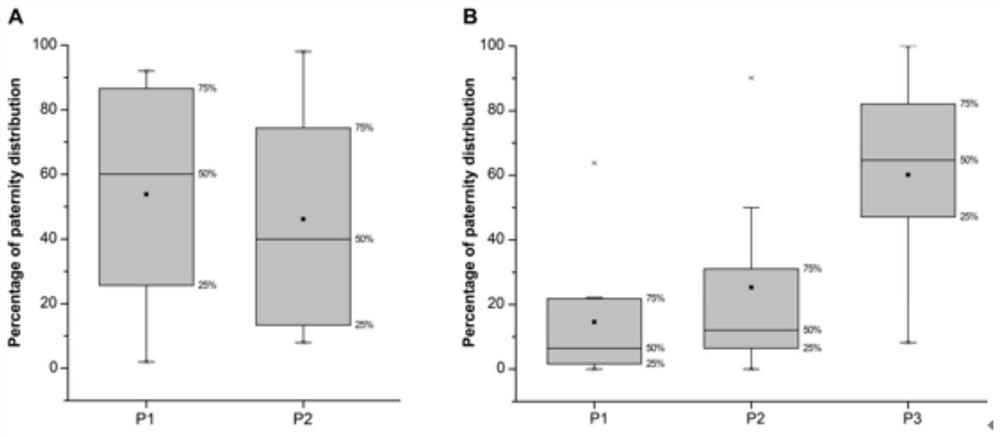
[0068] The identification of paternity of fireflies on the chest window
[0069] Select females with a body length of 24.20±2.78mm, a body width of 7.27±0.59mm, and a body weight of 0.34±0.09g, as well as a body length of 17.73±1.35mm, a body width of 6.78±0.59mm, and a body weight of 0.07±0.02g One female firefly was selected to mate with 2 male fireflies (n=16), and the other group was designed to mate with one female firefly and 3 male fireflies (n=9), and lay eggs and hatch into larvae, respectively. Each female and corresponding mating males and all successfully hatched progeny larvae were collected for SSR analysis, using fluorescently labeled PCR primers to amplify multiple microsatellite polymorphic sites, and then using high-resolution gel electrophoresis. The amplified fragments were separated, and the length of the amplified fragments was determined by a fluorescence detection system (Licor 4300) to realize the typing of the microsatellite polymorphic sites. Using ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com