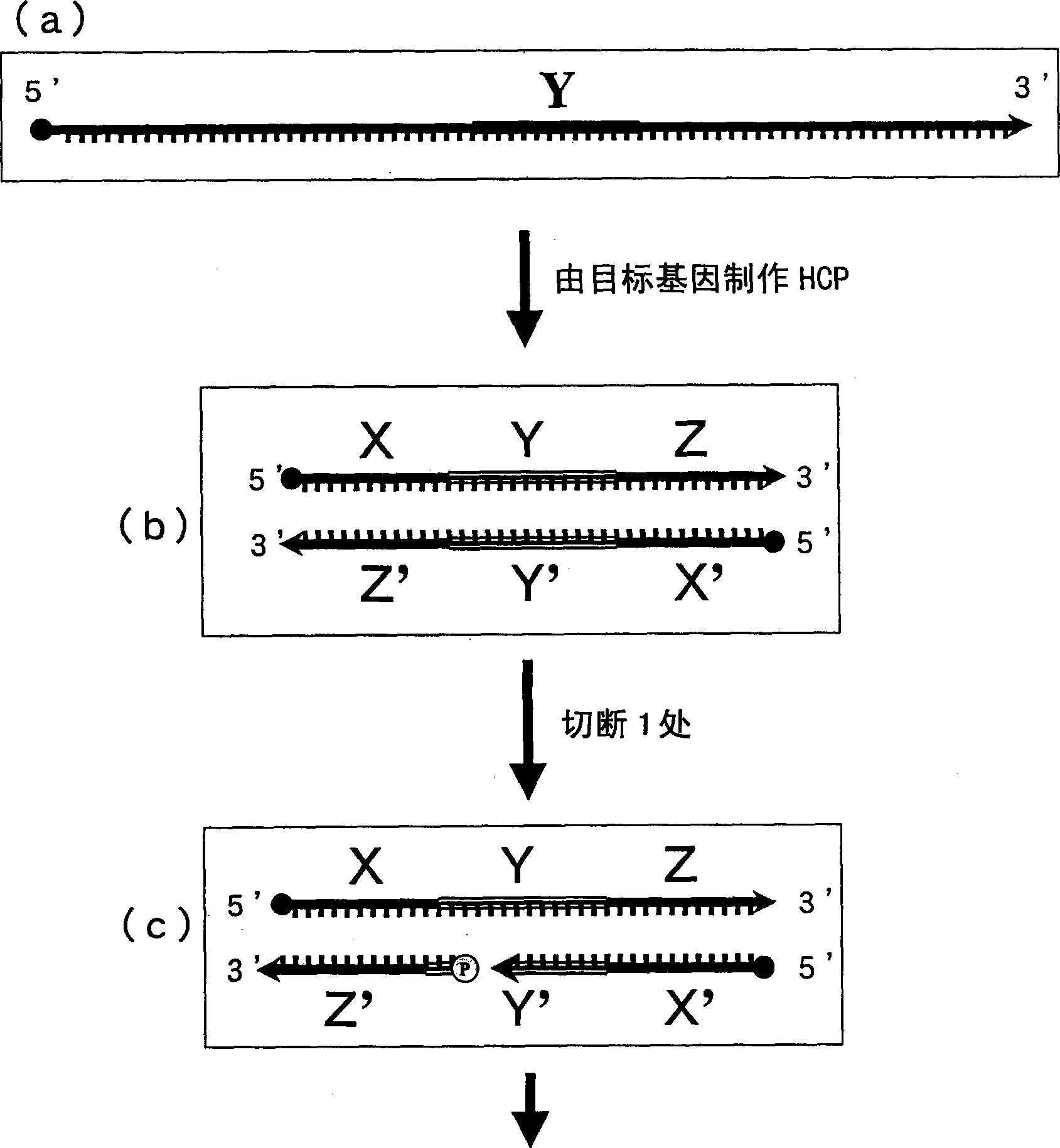
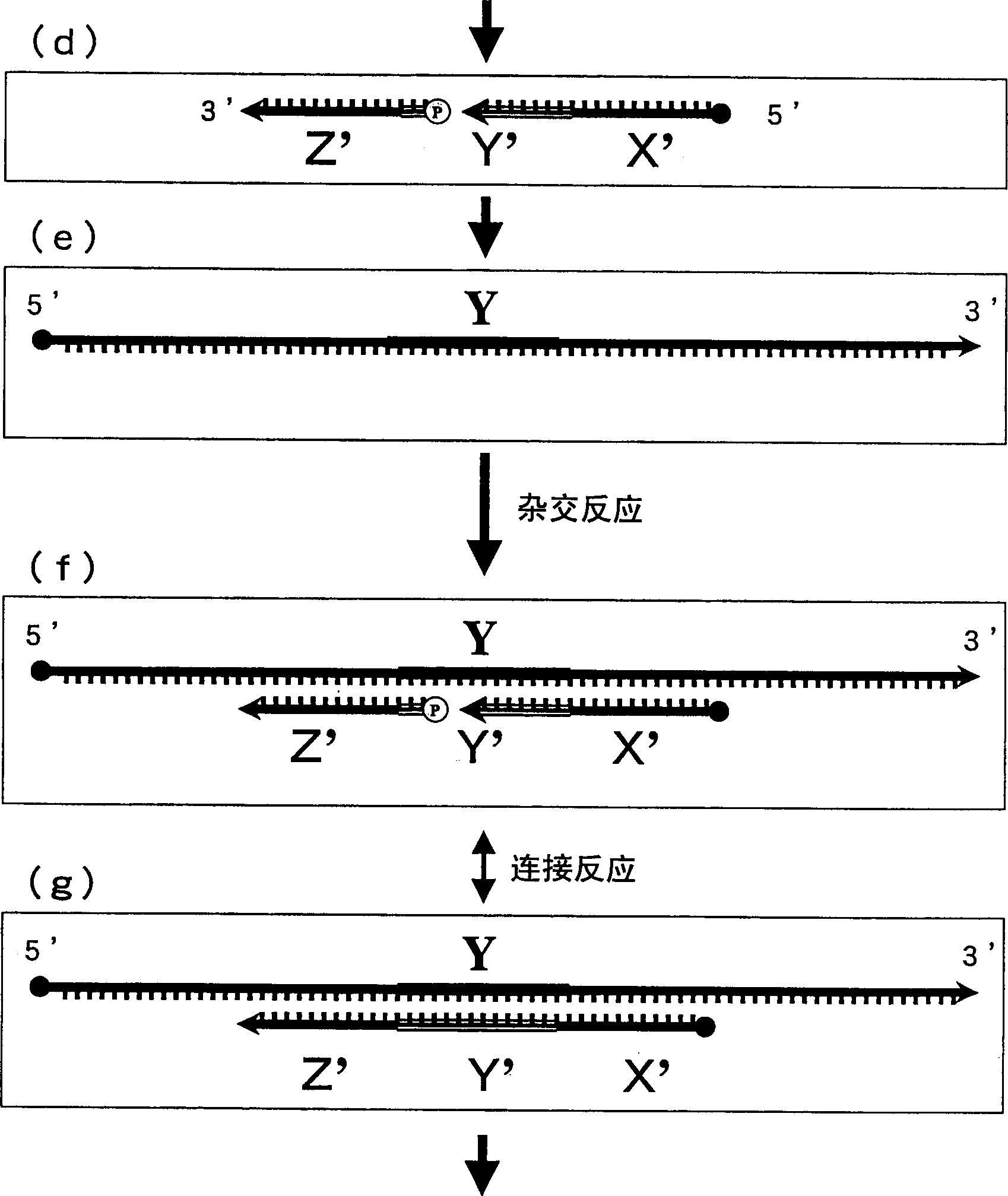
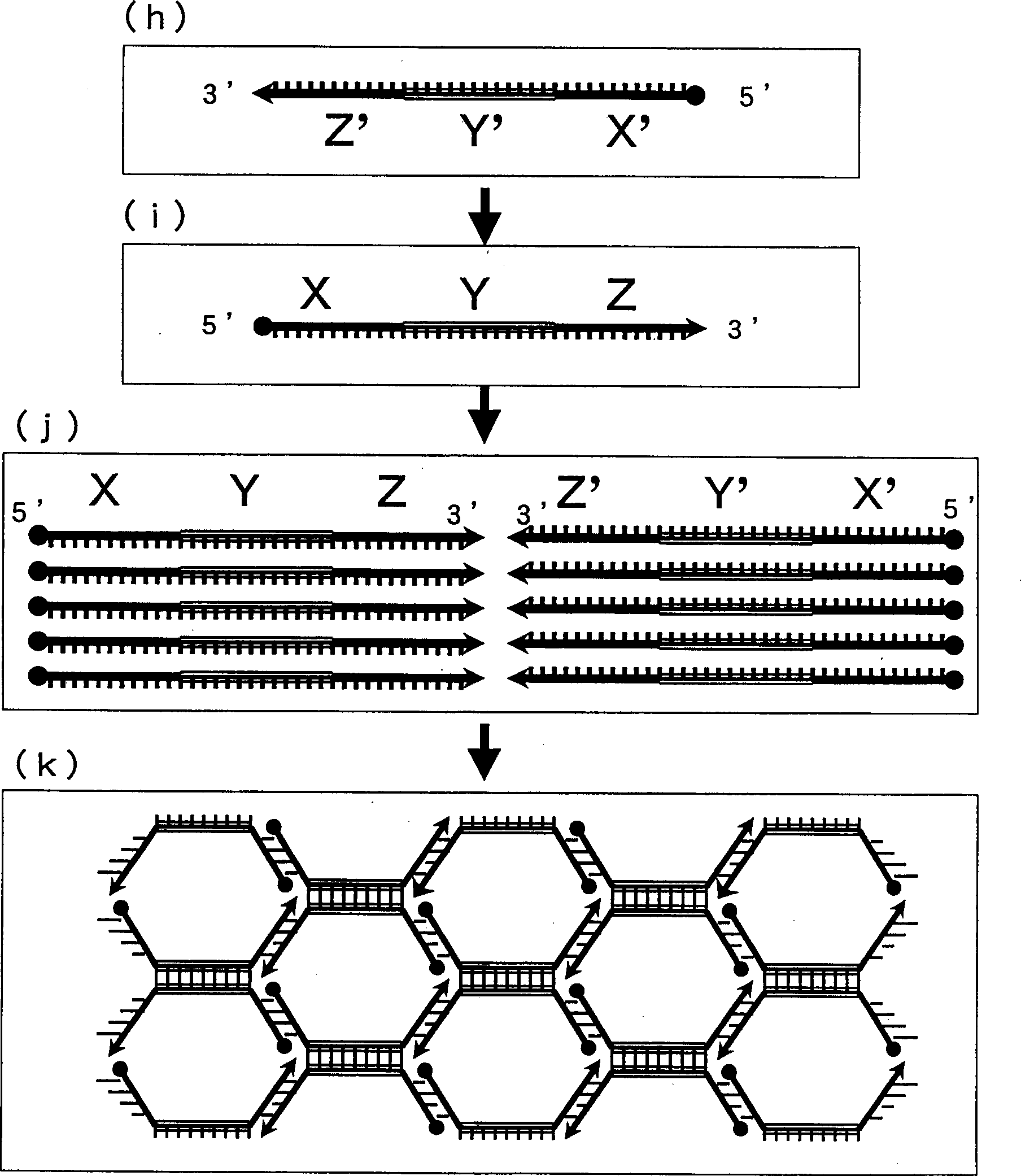
Method of detecting gene
A gene detection and target gene technology, which is applied in biochemical equipment and methods, microbial determination/inspection, fermentation, etc., and can solve problems such as inability to perform gene detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0079] (Example 1, Comparative Example 1 and Comparative Example 2) Ligation reaction in HCP (target gene: double strand)
[0080] 1. Purpose
[0081] Attempt to detect double-stranded target gene by ligation reaction of 1 cut HCP.
[0082] 2. Materials
[0083] 1) Regarding the target gene, a 60-base synthetic oligonucleotide derived from the Mycobacterium intracellulare 16s rRNA gene (hereinafter referred to as INT-①) and a synthetic oligonucleotide complementary to the target gene INT-① were produced (hereinafter referred to as INT-①R), the two were double-stranded, and the following experiments were performed.
[0084] 2) Preparation of probe 1 (hereinafter referred to as INT-1) having a region complementary to the Y region of the target gene INT-① and probe 2 (hereinafter referred to as INT-2) each region complementary to this probe 1 Constituted paired HCPs. Furthermore, probes obtained by cutting one of them (hereinafter respectively referred to as INT-1-A, INT-1-B,...
Embodiment 2~4 and comparative example 3
[0101] (Examples 2-4 and Comparative Example 3) Detection of SNPs by HCP ligation reaction
[0102] 1. Purpose
[0103] Discuss whether single-stranded target genes and SNPs can be detected by the ligation reaction of 1-cut HCP.
[0104] 2. Materials
[0105] In addition to the probe, ligase, ligation buffer, buffer and target gene INT-① used in Example 1, the target gene INT-①S1 (with the Y region of INT-① only 1 base different) was recreated. 1 base different from the part complementary to the 5' end of the Y region of HCP) and INT-①S2 (1 base different from the part complementary to the 3' end of the Y region of HCP).
[0106] 3. Method
[0107] a) Preparation of reaction solution
[0108] Add 0.5 μL of INT-1-A, INT-1-B, INT-2-C, and INT-2-D prepared at 100 pmol / μL respectively into 0.2 mL sterilized microtubes, and add 1 μL of ligase , culture buffer 2 μL, add 0.5 μL of the target gene prepared to 100 pmol / μL, pass DW 2 Prepare 20 μL of reaction solution respectively...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com