Method and cell for detecting modulations of RGS proteins
A protein, RGS4 technology, applied in cells modified by introducing foreign genetic material, biochemical equipment and methods, fusion cells, etc., can solve problems such as low affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
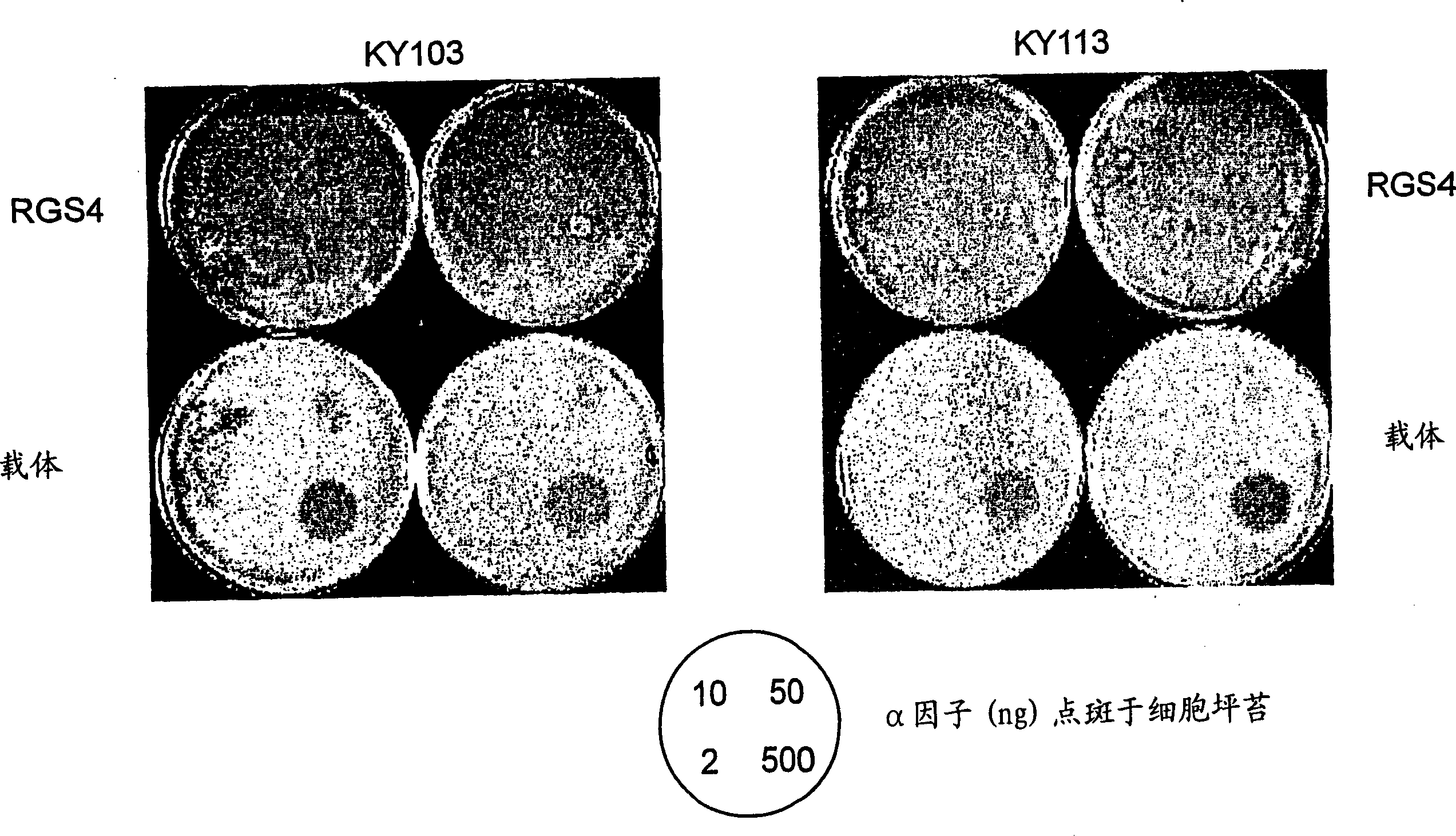
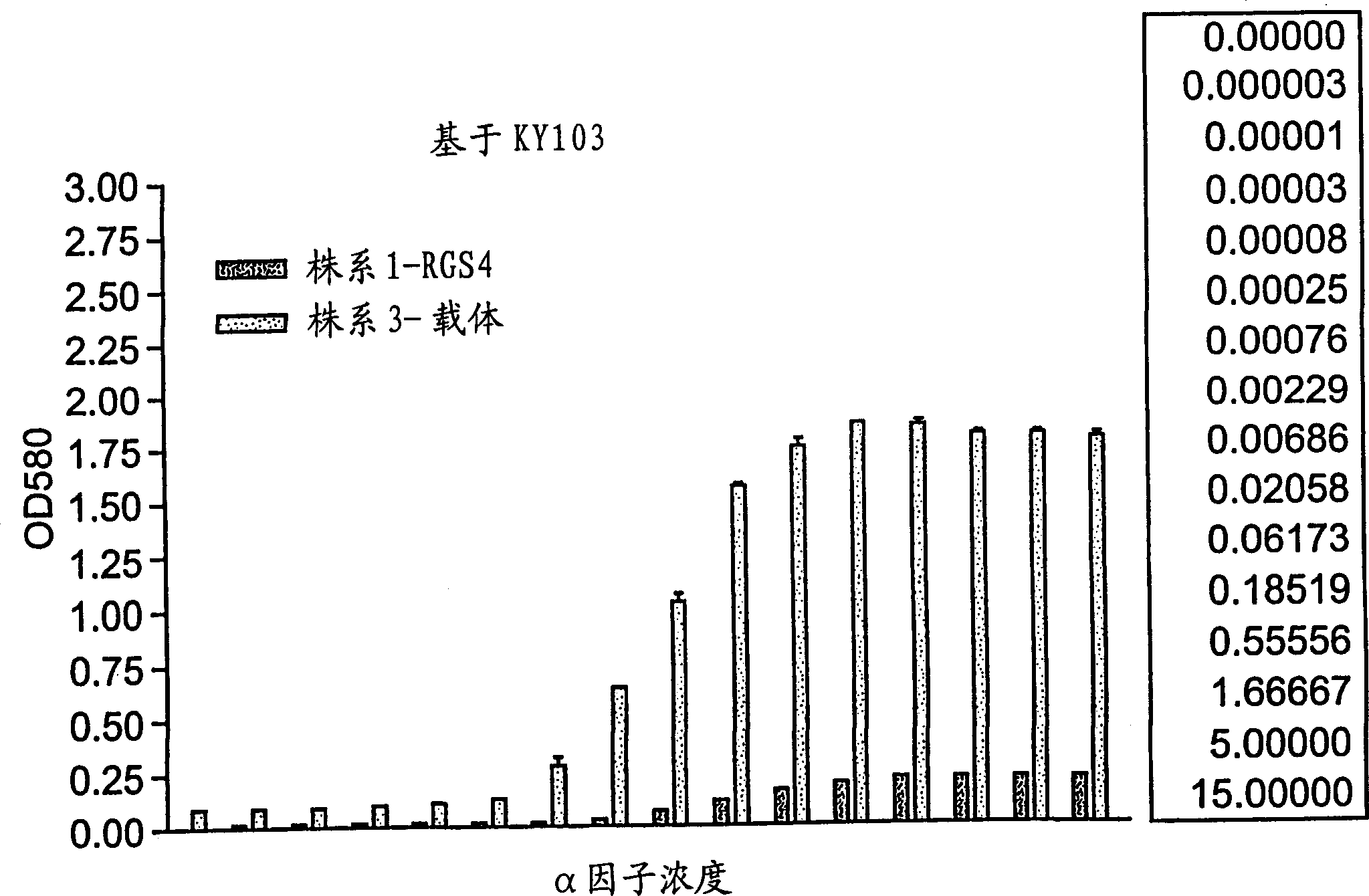
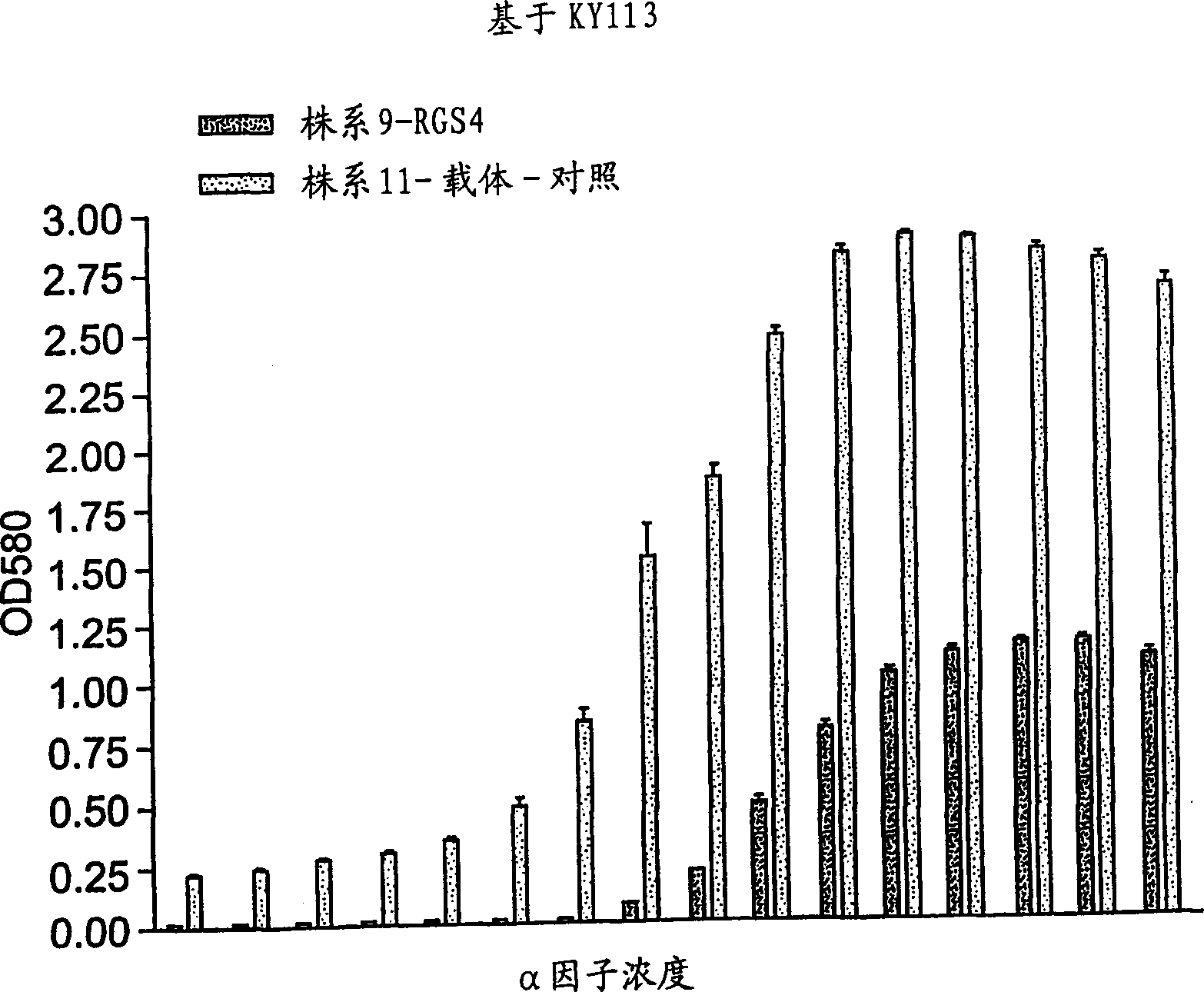
[0077] Example 1 RGS determination of FUS1-LacZ reporter gene:
[0078] GPCR receptor stimulation by alpha factor in yeast has been well characterized and characterized (see, Dohlman et al., 1996). Stimulation of this pathway normally causes cell cycle arrest in yeast. This signaling pathway is controlled by the endogenous RGS protein, Sst2, which functions as a negative regulator of GPCR signaling. The RGS protein acts to accelerate the endogenous GTPVase activity of the Gα subunit. Because deletion of the gene encoding Sst2 in yeast impairs its ability to turn off GPCR signaling, it causes hypersensitivity of the yeast cells to the ligand. The mammalian RGS protein, RGS4, is functionally complementary to the yeast Sst2.
[0079] In this experiment, reporter gene activity was measured in yeast strains lacking sst2 in the presence or absence of mammalian RGS4 protein. In this assay, we tested whether the RGS4 protein could regulate the alpha factor-induced pheromone respon...
Embodiment 2
[0102] Example 2 RGS assay of FUS1-luciferase reporter gene
[0103] Example 1 demonstrates that plasmids generated in response to the FUS1 promoter with the desired backbone and a pheromone operably linked upstream of the LacZ gene can be used to study the activity of RGS proteins. In this experiment, the luciferase gene was linked to the FUS1 promoter, not downstream of the LacZ gene. The luciferase system has several advantages over systems using the LacZ reporter, most notably the increased speed and ease of monitoring gene expression.
[0104] A FUS-1 reporter gene plasmid (Kp120) containing the luciferase gene was constructed. The FUS1 -luciferase reporter gene cassette was constructed by NcoI-XbaI digestion of plasmid pGL (Promega) to isolate the 1.7 kb fragment containing the firefly luciferase gene. This fragment was blunt-ended and purified. The Kp27 vector was used as the base vector, which was prepared by digestion with BamHI-NotI (to remove the LacZ gene), deph...
Embodiment 3
[0112] Example 3 Low copy version of the FUS1 luciferase reporter:
[0113] As a 2 [mu]m alternative to the aforementioned FUS1 luciferase reporter gene construct, a low copy number (CEN) version of the luciferase construct of Example 2 was prepared. A CEN version of the pheromone pathway responsive FUS1 luciferase reporter gene was generated by digesting plasmid Kp120 with KpnI and SacIEN to isolate a 2.8 kb fragment containing the FUS1 promoter and luciferase gene reporter cassette. Plasmids pRS414 and pRS416 (Stragagene) were digested with KpnI and SacI, respectively, and gel purified. Standard ligation was performed to generate two additional FUS1 luciferase recombinant plasmids; Kp133 (TRP1-tagged) and Kp135 / Kp131 * (URA3 tagged). Bacterial transformation and production of plasmid DNAs are performed by standard methods. Plasmids were confirmed by DNA sequencing. Plasmid Kp131 * Represents a similar construct for Kp135, but which was independently generated using a diff...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com