Method for detecting sequence of double chain DNA based on procedure of DNA automaton
A technology of DNA sequence and automaton, applied in the field of DNA sequence determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
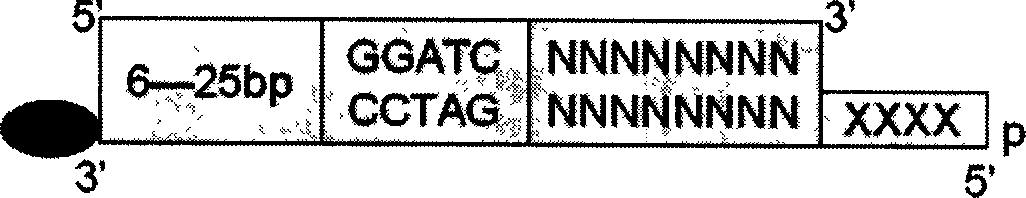
[0027] Determine the DNA sequence information of the unknown sequence that has been inserted into the multiple cloning site of the plasmid. Using DNA recombination technology to insert the sequence to be tested into the plasmid, the multiple cloning site should contain a recognition site for a restriction endonuclease such as FokI for the DNA calculation reaction; the known sequence of the multiple cloning site is the start of the determination sequence. The state transition molecule is designed so that the cut distance of each DNA automaton is 1 base, and the sequence of the DNA input molecule can be read directly. If FokI is used to run the automaton, the exposed cohesive ends are 4 bases, so the number of all state transition molecules that need to be considered is 4 4 = 256 pieces. It is only necessary to know the first 3 bases of the determined sequence (the known sequence of the multiple cloning site) to infer all subsequent sequences according to the fluorescent color...
Embodiment 2
[0052] Determine the sequence of the double-stranded DNA of the PCR product. It is necessary to design a FokI site in the PCR primer, so that one end of the double-stranded DNA molecule that is amplified by PCR (the specific method of the PCR reaction is not limited) contains a FokI site, which is far away from the end of the double-stranded DNA molecule. There is a distance of 6-10 bases at the end to ensure that this site can be effectively cleaved by FokI. In this way, the sequence determination of PCR products can be realized by using the DNA calculation reaction system corresponding to FokI. Before the reaction, the double-stranded DNA sequence amplified by PCR was cut with FokI to determine whether the sequence contained another FokI recognition site. If there is no other FokI recognition site, then directly perform the sequencing reaction, that is, the entire PCR product double-stranded DNA is added to the DNA calculation reaction system as an input molecule for DNA ca...
Embodiment 3
[0054] For unknown sequences that already contain a FokI recognition site, the following strategy can be used. The sequence was cleaved with FokI, and the cleaved fragments were recovered and directly subjected to a sequencing reaction. There are many ways to recover the fragments. The products of the FokI cleavage reaction can be separated by agarose gel according to the size of the fragments, and each fragment can be purified by tapping the gel separately. These recovery and purification processes utilize standard DNA recombination techniques, and any kits on the market can be selected to carry out. The purified DNA fragments can be directly subjected to the DNA calculation reaction to determine the sequence, that is, the purified DNA double-stranded fragment is added to the DNA calculation reaction system as the input molecule of the DNA calculation. Finally, the sequences of these fragments are spliced together.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com