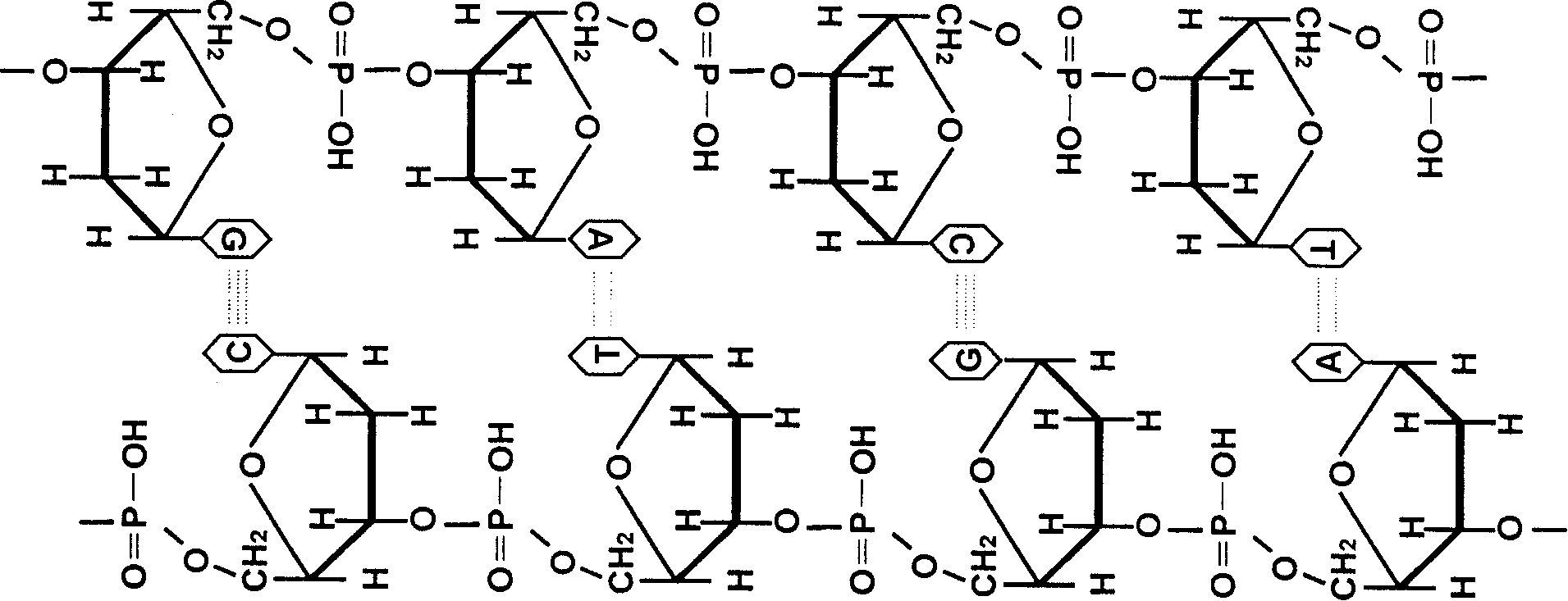
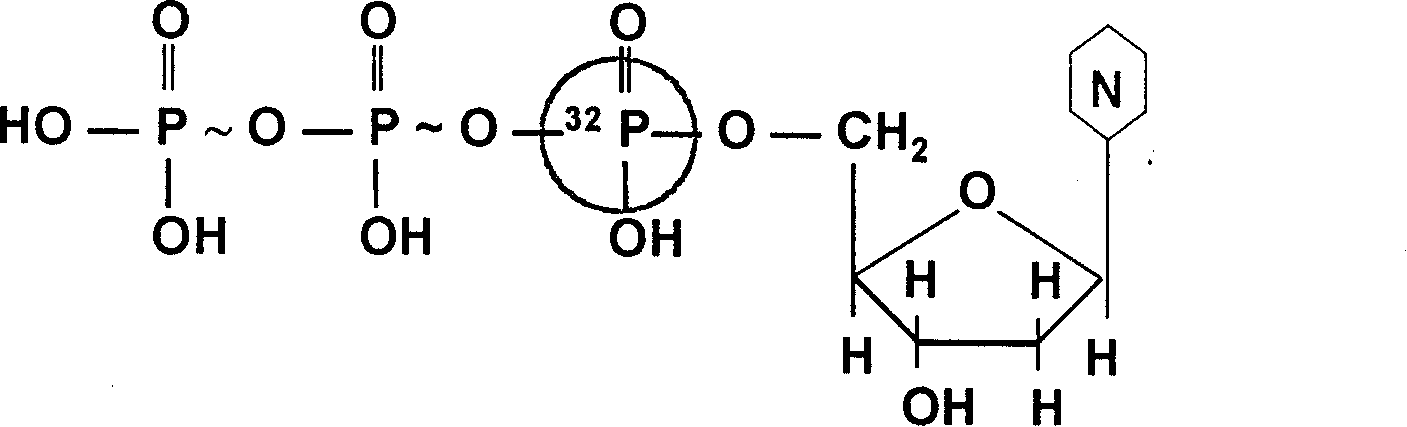
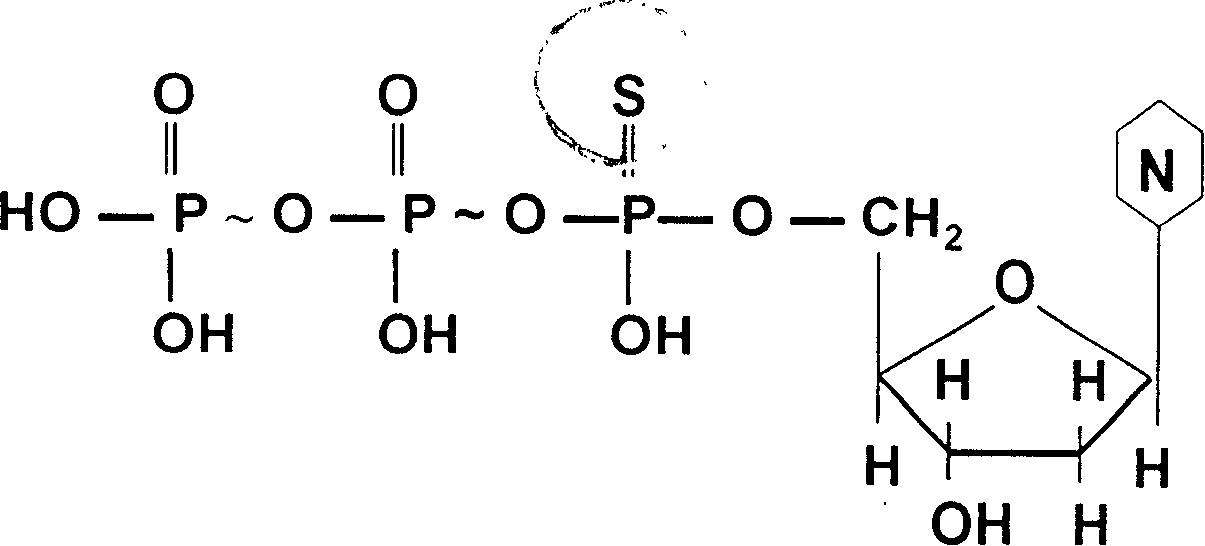
Method for deriving mulriple DNA sequences from atom destructurizing
A technology of DNA sequence and diversity, applied in the field of gene mutagenesis research, can solve problems such as low degree of mutation, multiple types of mutation, and carcinogenic risk of chemical mutagens, achieving high biological safety, multiple types, and high tendency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Using the onion Ace-AMP1 gene (357bp) as a template to introduce α- 32 P-dATP
[0031] ①Primer design: The onion Ace-AMP1 gene was cloned on the pUC19 plasmid vector, and PCR primers were designed according to its sequence—the upstream primer was 5’-TAA GGT A CC ATG G TTCGC GTT GTA TC-3' (including restriction site NcoI), the downstream primer is 5'-TCC TGCCGC ATT GAA TTC TTG G-3' (including restriction enzyme site EcoRI), the theoretically amplified DNA fragment length is 357bp;
[0032] ②α- 32 The introduction of P-dATP (i.e. the first round of PCR): with the source DNA in step ① as a template, α- 32 P-dATP makes α- 32P-dATP: normal dATP=1:15, 35 cycles at 94°C for 1 min, 55°C for 0.5 min, and 72°C for 1 min to complete the first round of PCR reaction;
[0033] ③Radioactive decay: place the PCR amplification product in step ② at -20°C to decay for 7 half-lives;
[0034] ④The second round of PCR: use the decayed PCR product in step ③ as the template...
Embodiment 2
[0037] Embodiment 2: Using the specific DNA sequence (301bp) of wheat as a template, the introduction of α- 32 P-dATP
[0038] ①Primer design: clone a specific DNA sequence from wheat into the pGEM-T plasmid vector, and design PCR primers according to its sequence—forward primer E-AG is 5'-CCCGGG CAG GTA ATT CAG-3', complementary The strand primer M-CG is 5'-TCG CGG CCG AGG TTAACA-3', the theoretically amplified DNA fragment length is 301bp;
[0039] ②α- 32 The introduction of P-dATP (i.e. the first round of PCR): with the source DNA in step ① as a template, α- 32 P-dATP makes α- 32 P-dATP: normal dATP=1:15, 35 cycles at 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min to complete the first round of PCR reaction;
[0040] ③Radioactive decay: place the PCR amplification product in step ② at -20°C to decay for 7 half-lives;
[0041] ④The second round of PCR: use the decayed PCR product in step ③ as the template, and use the normal dNTP as the reaction substrate to perform...
Embodiment 3
[0044] Example 3: Using the specific DNA sequence (301bp) of wheat as a template to introduce α- 32 P-dCTP
[0045] What is incorporated in the reaction substrate dNTP in step ② is α- 32 For P-dCTP, other steps are the same as in Example 2; step ⑥ sequencing and analysis: 10 clones in step ⑤ are randomly selected for sequencing analysis by Songbao Biological Company. The results showed that: 9 of the 10 clones had mismatches at expected positions, of which: 1 had mismatches at 5 positions; 1 had mismatches at 4 positions; 2 clones had mismatches at 2 different positions Mismatches and deletions of 1 base; 5 had 1 mismatch at a different position. Controls have no mismatches at any position. The positions where replication mismatch occurs are bases C and G, and the rule of mismatch is that C is mismatched to T, and G is mismatched to A, that is, base conversions of G→A and C→T occur (see Sequence Table 3).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com