Saliva streptococcus 57.I urase gene and its urase
A technology of Streptococcus salivarius and urease, applied in gene therapy, enzymes, enzymes, etc., can solve problems such as cloning that has not been reported yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Extraction and Identification of Genomic DNA of Streptococcus salivarius 57.I
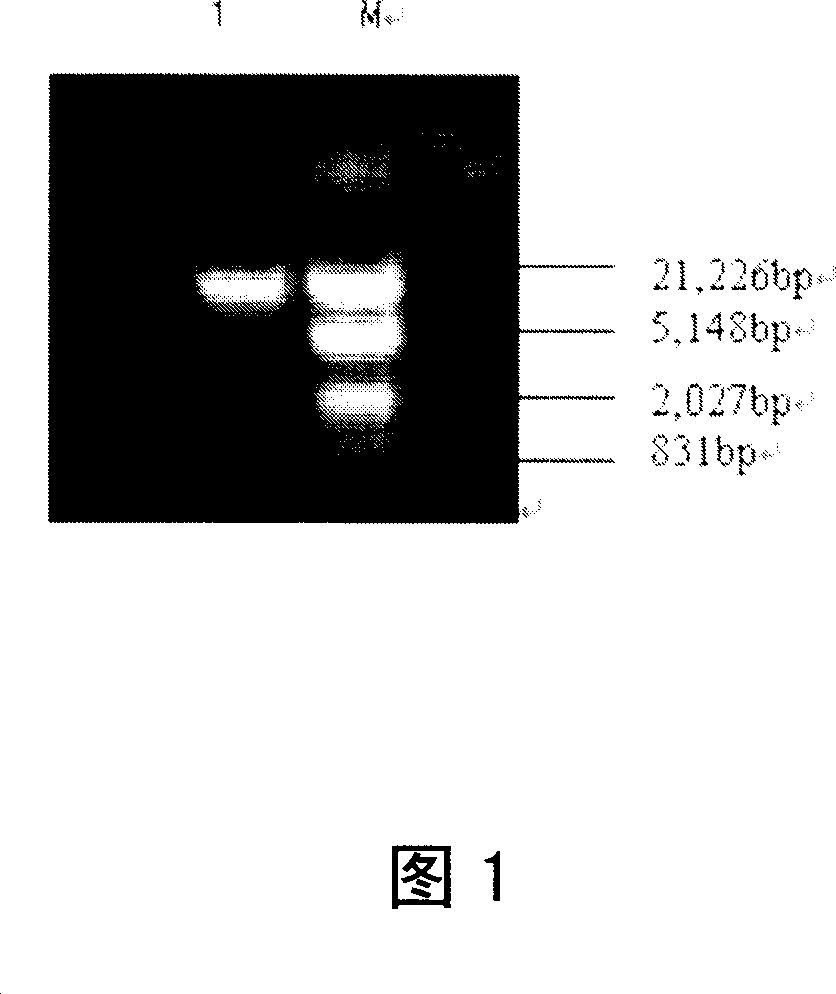
[0048] The purpose of the whole study is to obtain the clone of the active Streptococcus salivarius 57.I urease gene U35248. The first step is to obtain the template of the target gene, which is the whole genome DNA of the donor bacteria. Therefore, this experiment According to the principle of molecular cloning, the genomic DNA of Streptococcus salivarius 57.I will be extracted using a commercially available bacterial genome extraction kit as a template, so that in subsequent studies, PCR methods will be used to use genomic DNA as a template. The desired urease gene was cloned.
[0049] At the same time, because in the genome of prokaryotes, the DNA molecule encoding 16S ribosomal RNA is functionally stable and consists of a highly conserved region and a variable region. The mutation rate in the conserved region is extremely low, while the variable sequence of its characteristic region is d...
Embodiment 2
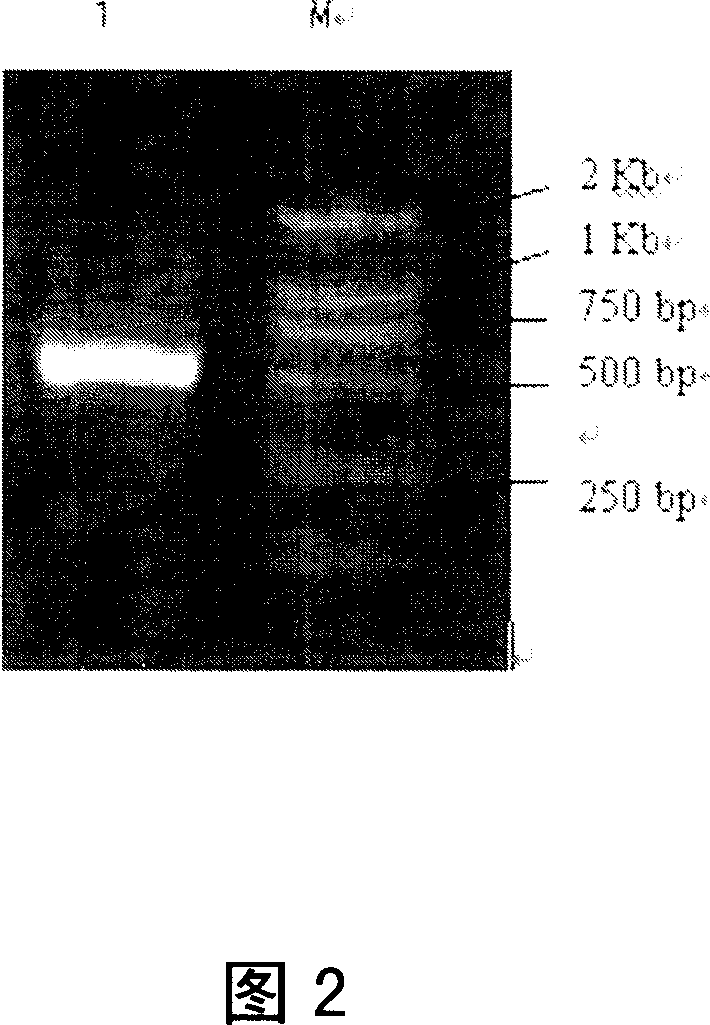
[0117] Segmental Cloning of 57.I Urease Gene U35248 from Streptococcus salivarius
[0118] Polymerase chain reaction (polymerase chain reaction, PCR) can be used to amplify the DNA segment between two pieces of known sequences, but the length of Streptococcus salivarius urease gene reaches 8Kb, if PCR method is directly used for one-time Amplification, it is difficult to ensure that the expected length can be achieved, and more mutations may appear during the amplification process. This experiment intends to adopt the method of segmental cloning. After analyzing the full-length sequence of Streptococcus salivarius urease gene U35248, it is found that there are two single restriction endonuclease cutting sites-Bam H I inside the gene. (located at 2038bp) and Xho I (located at 4812bp), so the design divides the urease gene into three sections, front, middle and back, and clones them respectively. There is a part of overlap between every adjacent two sections, and each The overl...
Embodiment 3
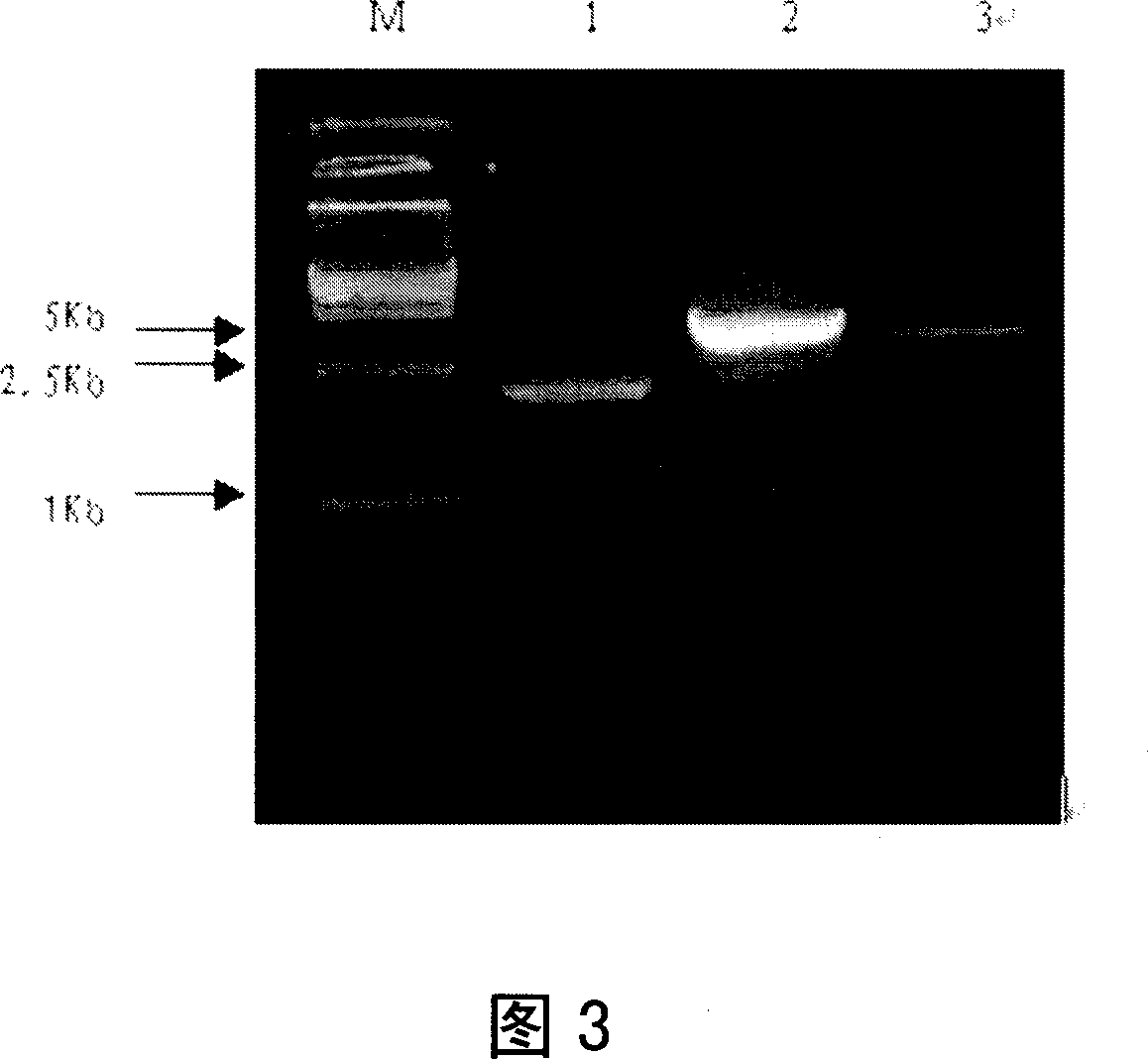
[0253] The products of segmental cloning were connected step by step into a complete urease gene.
[0254] When initially segmenting the urease gene U35248 of Streptococcus salivarius 57.I, there was an overlapping region between the first segment and the second segment and the second segment and the third segment of the target gene segment. Each has a single enzyme cutting site. In the above experiments, the three fragments have been connected into the same vector in the same direction. In this experiment, a single restriction site on the vector and a single restriction site inside the fragment will be used, and the method of double digestion will be used to target The gene and the vector were cut and then recombined, so that the three segmented clones that had been correctly sequenced in Experiment 3 were gradually connected into a complete urease gene.
[0255] 1.1 Materials and methods
[0256] 1.1.1 Main reagents
[0257] A small amount of DNA fragment quick gel recove...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com