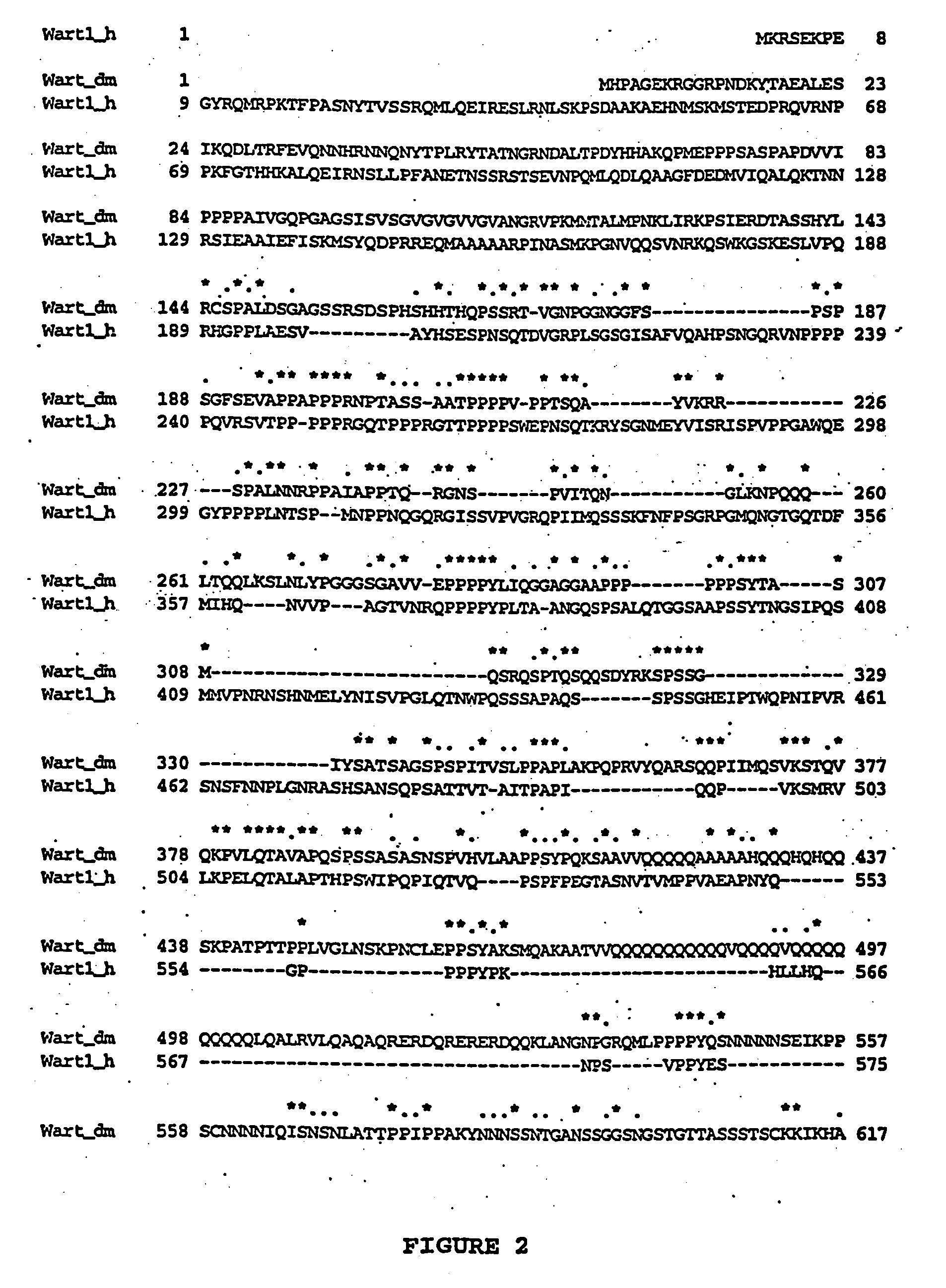
Human orthologues of WART
a technology of human orthologues and warts, which is applied in the field of protein kinase proteins, can solve the problems of growth of tumors on the legs and wings of flies, and achieve the effect of facilitating the uptake of compound
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning of Murine WART1
[0207] Total RNAs were isolated using the Guanidine Salts / Phenol extraction protocol of Chomczynski and Sacchi (P. Chomczynski and N. Sacchi, Anal. Biochem. 162:156, 1987) from murine embryos from gestational day 12. These RNA were used to generate single-stranded cDNA using the Superscript Preamplification System (GIBCO BRL, Gaithersburg, Md.; Gerard, G F et al., Focus 11:66, 1989). A typical reaction used 10 μg total RNA with 1.5 μg oligo(dT)12-18 in a reaction volume of 60 μL. The product was treated with RNaseH and diluted to 100 μL with H2O. For subsequent PCR amplification, 1-4 μL of the sscDNA was used in each reaction.
[0208] Degenerate oligonucleotides targeted for the Epidermal Growth Factor (EGF) family were synthesized on an Applied Biosystems 3948 DNA synthesizer using established phosphoramidite chemistry, precipitated with ethanol and used unpurified for PCR. The sequence of the degenerate oligonucleotide primers used were the following:
(SEQ ...
example 2
cDNA Cloning and Characterization of Human WART1
[0212] A second PCR strategy was designed to isolate the human orthologue of the novel mouse clone. Degenerate primers based on clone 105-4-10 were used to amplify templates derived from a pool of primary human non-small cell lung carcinomas. Total RNAs from primary human lung tumors were isolated as in Example 1. The sequence of the degenerate oligonucleotide primers used were as follows:
(SEQ ID NO:9)5774 = 5′ - TCCRAACAGDATNACNCCNACNSWCCA - 3′and(SEQ ID NO:10)5326 = 5′ - TTYGGNYTNTGYACNGGNTTYMGNTGG - 3′.
These primers were derived from the sense and antisense strands, respectively of peptide sequences FGLCTGFRW (SEQ ID NO:11) and WSVGVILFE (SEQ ID NO:12) present in the murine WART1 clone. The amplification conditions were similar to those described in Example 1 using oligonucleotides KITDFG (SEQ ID NO:7 and KCWMID (SEQ ID NO:8). Two distinct PCR products were isolated, SuSTK15 (268 bp) and SuSTK17 (273 bp). These two fragments sha...
example 3
Isolation of hWART1
[0213] A human bone marrow gt11 cDNA library was probed with the PCR fragments corresponding to human WART1. Probes were 32P-labeled by random priming and used at 2×106 cpm / mL following standard techniques known in the art for library screening. Prehybridization (3 h) and hybridization (overnight) were conducted at 42° C. in 5×SSC, 5×Denhart's solution, 2.5% dextran sulfate, 50 mM Na2PO4 [pH 7.0], 50% formamide with 100 mg / mL denatured salmon sperm DNA. Stringent washes were performed at 65° C. in 0.1× SSC and 0.1% SDS. DNA sequencing was carried out on both strains using a cycle sequencing dye-terminator kit with AmpliTaq DNA Polymerase, FS (ABI, Foster City, Calif.). Sequencing reaction products were run on an ABI Prism 377 DNA Sequencer.
[0214] Three cDNAs were isolated and completely sequenced. Two of the clones were found to be overlapping clones that encoded a long C-terminal open reading frame (ORF) but lacked an upstream stop codon. The third clone was fo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap