Micrornas expression signature for determination of tumors origin
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Specific microRNAs are Able to Distinguish Between Primary Non-Hepatic and Hepatic Tumors
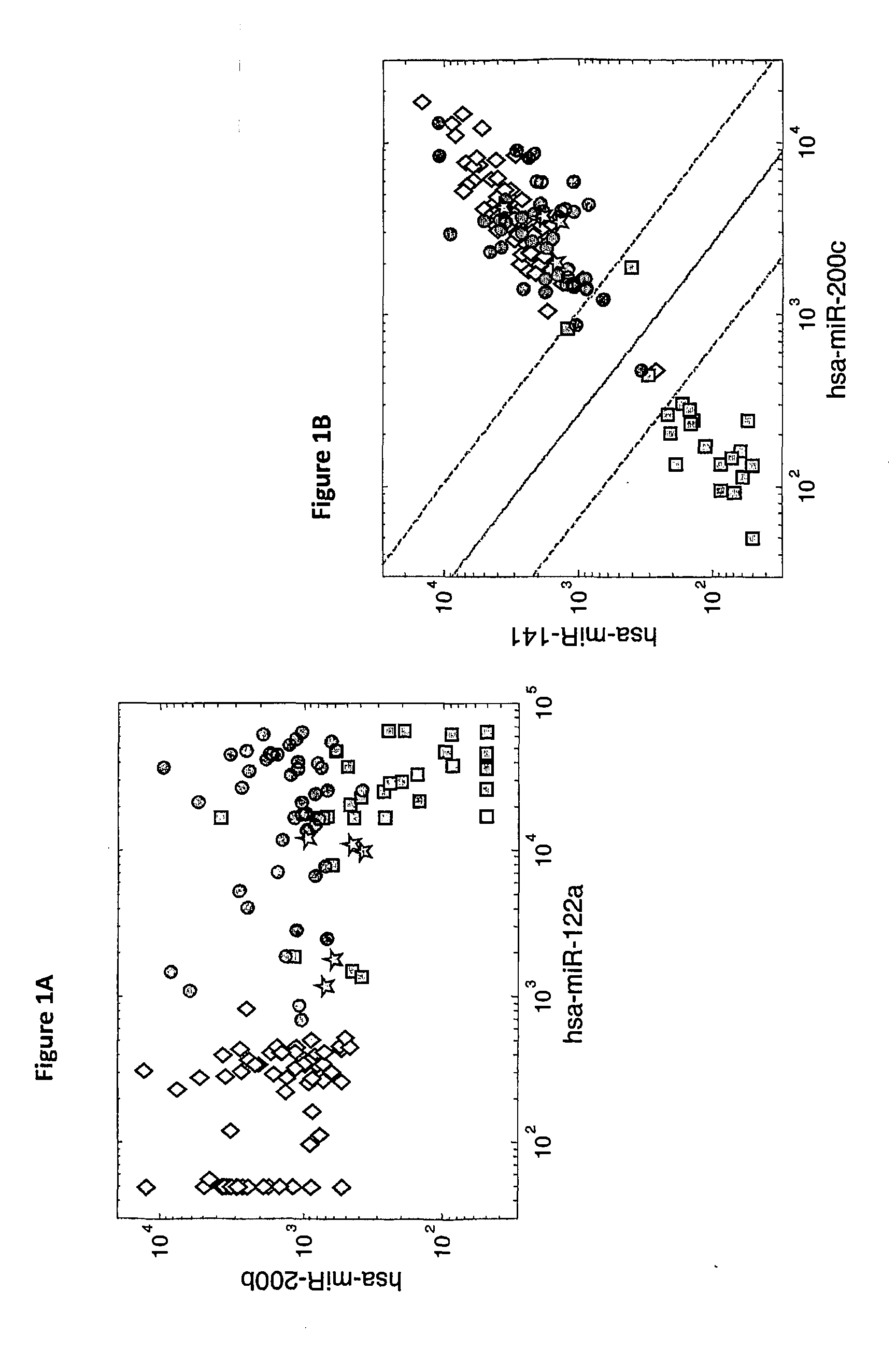
[0205]microRNA expression levels were profiled in 144 tumor samples including 30 primary HCC samples, 63 primary tumors from epithelial origins, 46 liver metastases from epithelial origins, and 5 adenocarcinoma metastases to the liver from unknown origin. The primary HCC samples were compared to the other primary tumors and to the liver metastases samples. Hsa-miR-122a (SEQ ID NO: 32), which is a highly liver-specific microRNA, had the strongest effect when comparing primary HCC tumors to other primary tumors with a fold-change>90, and could identify HCC from other primary tumors (p-value=1.4e-38, AUC=1). However, this microRNA is also found at high levels in the RNA extracted from liver metastases (FIG. 1A), ostensibly due to contamination from the adjacent normal liver tissue, and is not a good marker for identifying liver metastases (fold-change of medians 1.1, p-value=0.28, AUC=0.56). By usi...
example 2
Specific microRNAs are Able to Distinguish Between Primary GI Tumors and Non-GI Primary Tumors
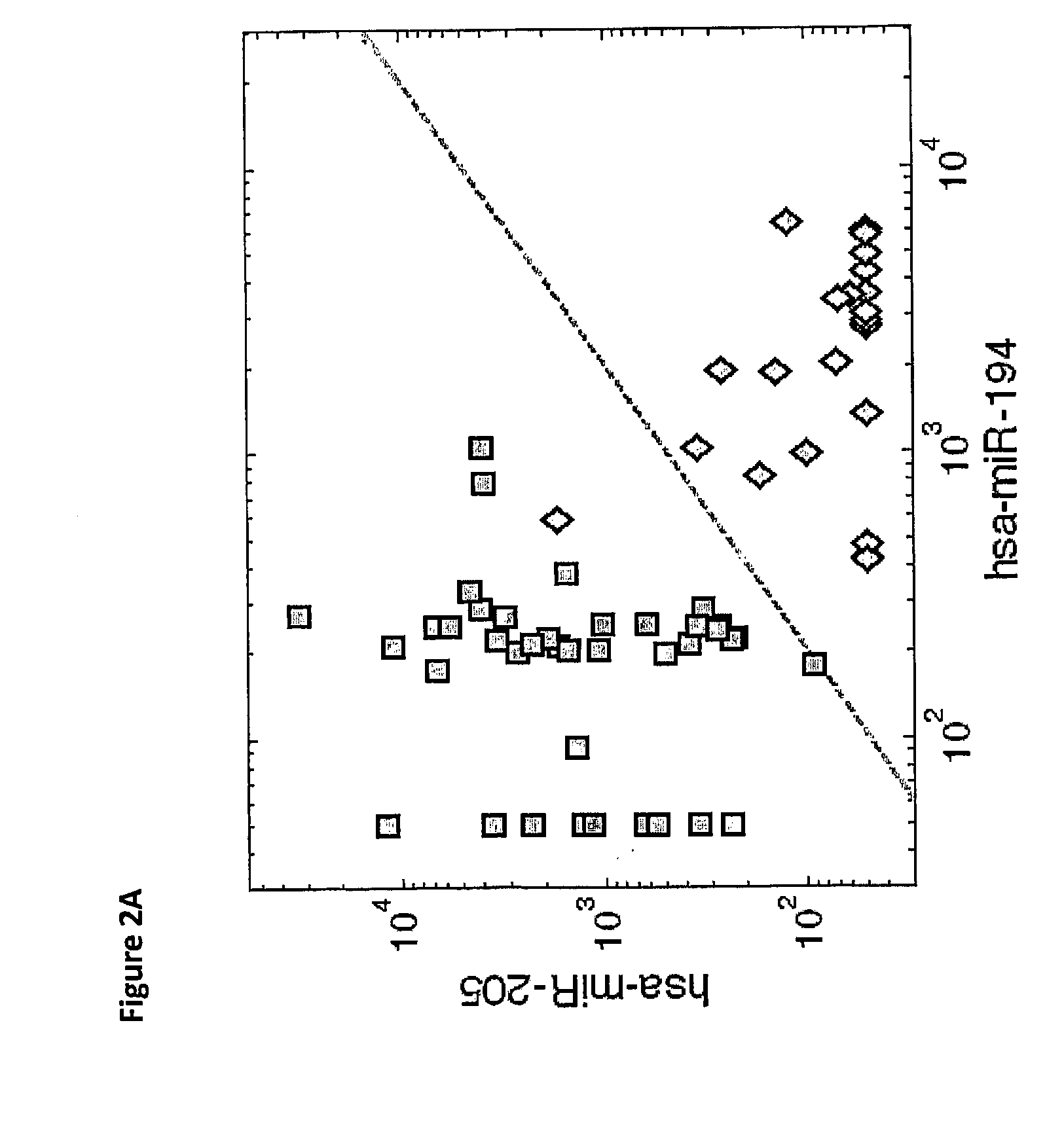
[0209]The microRNA expression can provide further information on the possible origin of liver metastases. Another pair of microRNAs, hsa-miR-194 (SEQ ID NO: 1) and hsa-miR-205 (SEQ ID NO: 4), had significant different expression (p-value<1e-12 for each) in primary tumors from gastrointestinal (GI) origin (14 colon, 5 pancreas, 5 stomach) compared to primary tumors of non-GI epithelial origin (24 lung, 15 breast). The ratio of these expression levels could be used to accurately identify primary tumors from non-GI origin: the decision rule “classify as non-GI primary when the expression of hsa-miR-205 is greater than half the expression of hsa-miR-194” (FIG. 2A; dashed line marks the decision boundary) had a sensitivity of 100% and specificity of 96% (AUC=0.9893). In the liver metastases, despite the small number of samples, this trend was maintained (FIG. 2B). However, since hsa-miR-194 is a...
example 3
Specific microRNAs are Able to Distinguish Between Primary Brain Tumors, Brain Metastases, Non-Brain Primary Tumors and Normal Brain Samples
[0210]microRNA expression levels were profiled on a microarray platform in 252 tumor samples including 15 brain primary tumor samples, 187 non-brain primary tumors, 50 brain metastases from various tissue origins and 2 normal brain samples. The brain primary tumor samples were compared to the other primary tumor samples, to normal brain samples and to samples of brain-located metastases (Table 6). Hsa-miR-124 (SEQ ID NO: 16), which is highly specific to the nervous system, displayed the greatest disparity in expression when comparing brain primary tumors to other primary tumors, with a fold-change of ˜100 (p-value=5.1e-57, AUC=0.9976, see Table 6). A combination of hsa-miR-124 and hsa-miR-219 (SEQ ID NO: 24) (B0, see methods) could be used to distinguish brain primary tumors from non-brain primary tumors with 100% accuracy (FIG. 4A). Other brain...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com