Protease inhibition
a protease and inhibitory technology, applied in the direction of enzyme inhibitors, peptide/protein ingredients, immunological disorders, etc., can solve the problems of a lack of specificity for particular adams, serious drawbacks of the methods which have been identified to date for inhibiting metalloproteases such as adams, and difficulty in predicting potential cleavage sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1a
Synthesis of Cyclic Peptides
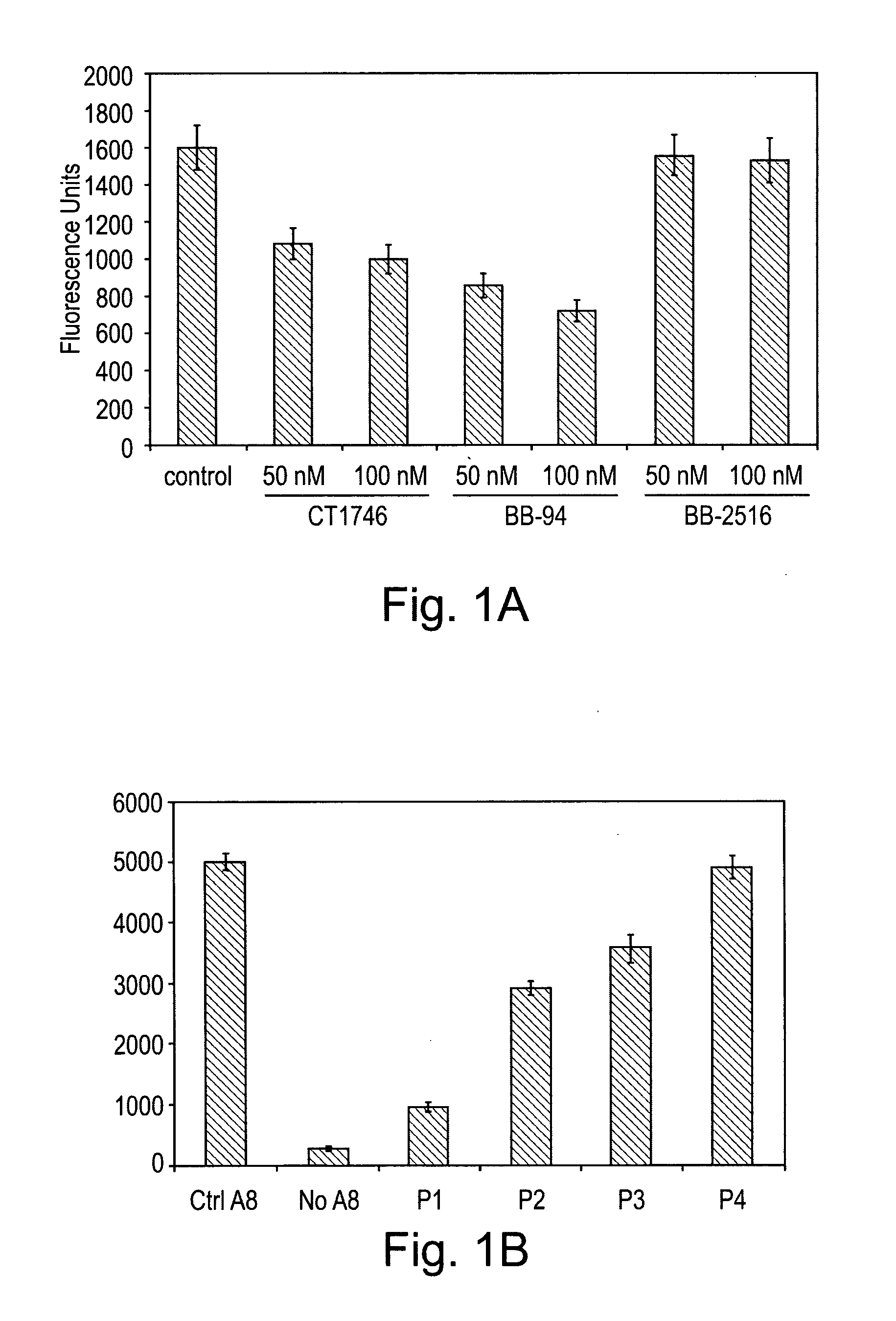
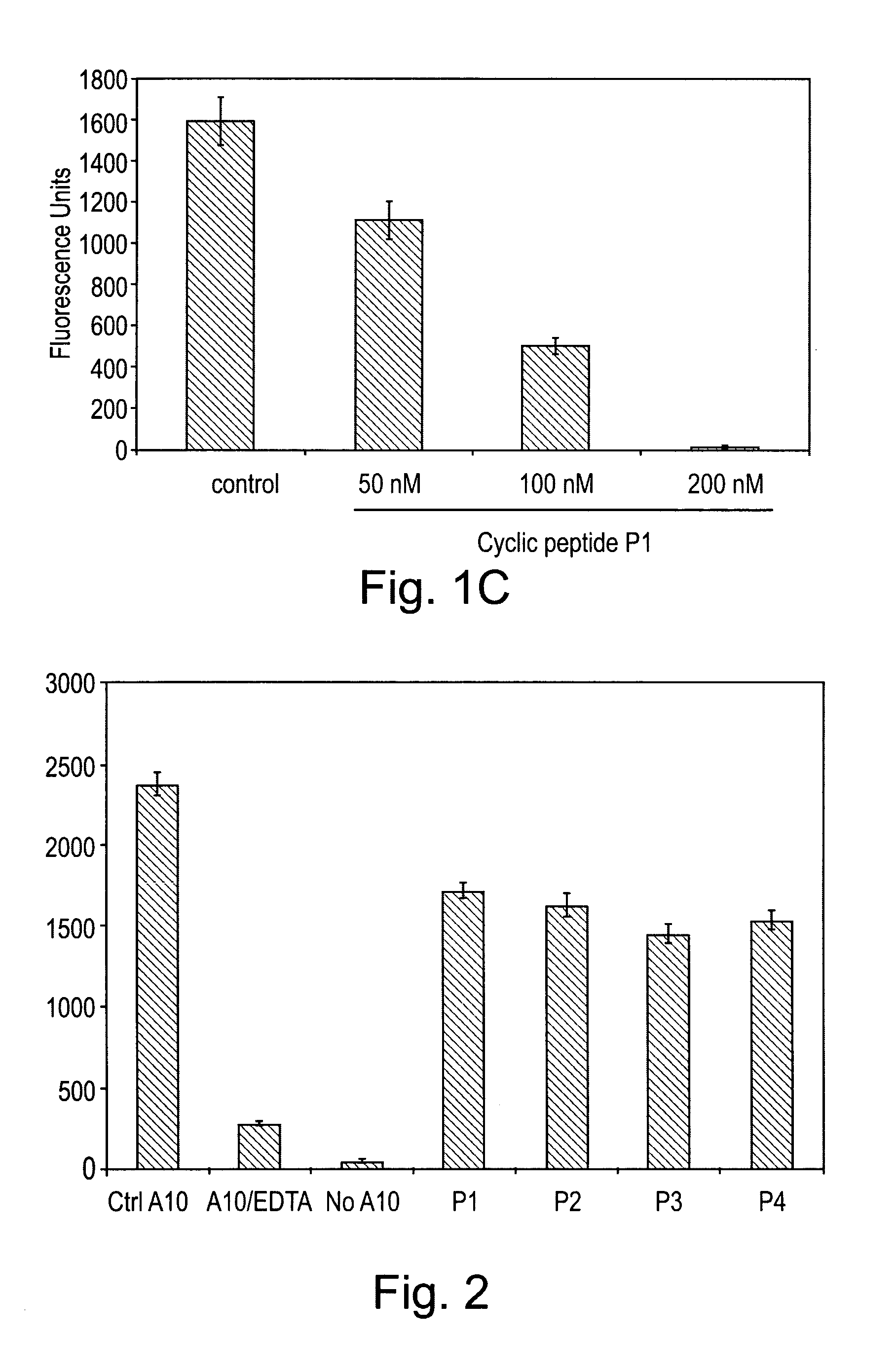
[0101]In the following example, a series of peptides were synthesised in order to test their activity as inhibitors of ADAM proteases. The structures of the peptides are shown in Table 1 and FIG. 5.
[0102]Each peptide comprises 6 amino acid residues. Peptides P1 to P4 have the sequence RLSKDK, which is found in the integrin binding loop of the disintegrin domain of murine ADAM8. The KDK sequence of ADAM8 is at an equivalent position to the RGD sequence found in the integrin-binding loop of ADAM15. Peptides P1 to P3 are cyclic peptides comprising this motif, whereas P4 is a linear peptide with the KDK sequence at the C terminal. Each of P1 to P3 comprises a single β-amino acid, the position of which differs between the three peptides.
[0103]Each of peptides P5, P6 and P7 is a cyclic peptide which differs in sequence from peptide P1 at one amino acid position within the KDK motif found in P1. Each of P5 to P7 comprises a single β-amino acid residue at the sam...
example 1b
Synthesis of Cyclic Peptides
[0106]The peptides described in Example 1 may also be synthesised using the following method:
[0107]Cyclic peptides such as P1 were synthesised according to Malesevic et al., 2004, published in Journal of Biotechnology 112, 73-77. Briefly, linear peptides were synthesized according to an Fmoc-protocol with Wang resin as a solid support. In a typical experiment, each peptide coupling is done twice with 1.5 eq. Fmoc-amino acid (0.3M in DMF*), 1.5 eq. TBTU (0.3M in DMF) and 3 eq. DIPEA (0.6M in DMF). After washing with DMF the Fmoc group is cleaved with a solution of 2% piperidine and 2% DBU in DMF. Wang resin is used as the solid support which is preloaded with Fmoc-Asp-ODmb attached to the resin via the side chain functionality. After assembly of the peptide sequence, the terminal protecting groups (Fmoc at the N-terminus, Dmb at the C-terminus) are cleaved from the linear precursor. A solution of 1.0-3.0 eq. (relative to resin loading) of HATU in 3 mL DMF ...
example 2
Inhibiting Cell Adhesion Mediated by ADAM8 DC Domain Using Cyclic Peptides
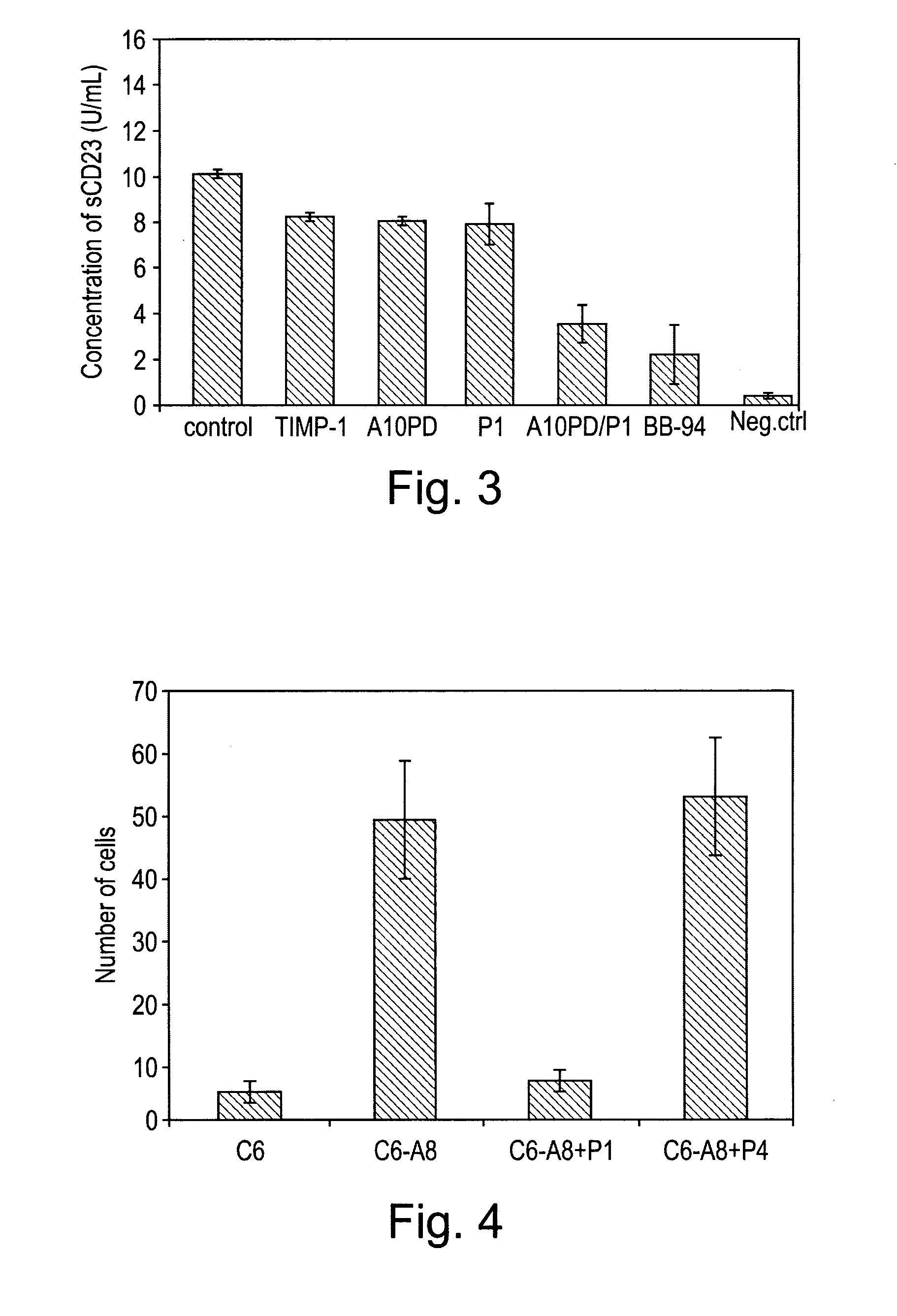
[0109]In this example, peptides synthesised as described in example 1 were used to target cell adhesion mediated by the ADAM8 DC domain. A recombinant DC domain of ADAM8 was expressed and purified from E. coli and then coated onto cell culture plates. Cells over-expressing ADAM8 were then allowed to bind to the ADAM8 DC domain on the plates.
[0110]96-well plates were coated with 50 μg / ml of recombinant ADAM8 DC protein in PBS at 4° C. for 16 h. To express the recombinant DC domain of ADAM8, the cDNA fragment encoding the A8 DC domain was generated by PCR using Platinum™ Pfx polymerase (Invitrogen) with the following primers: DCE-A8f, 5′-GGT GGC CCT GTG TGT GGA AAC-3′ (SEQ ID NO:17); DCE-A8r, 5′-TAC ACA GTT GGG TGG TGC CCA-3′ (SEQ ID NO:18). The resulting cDNA fragment was cloned into the bacterial expression vector pTrcHis2 (Invitrogen) containing a C-terminal Myc and His6 tag. This vector was transformed into ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Structure | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
| Pharmaceutically acceptable | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com