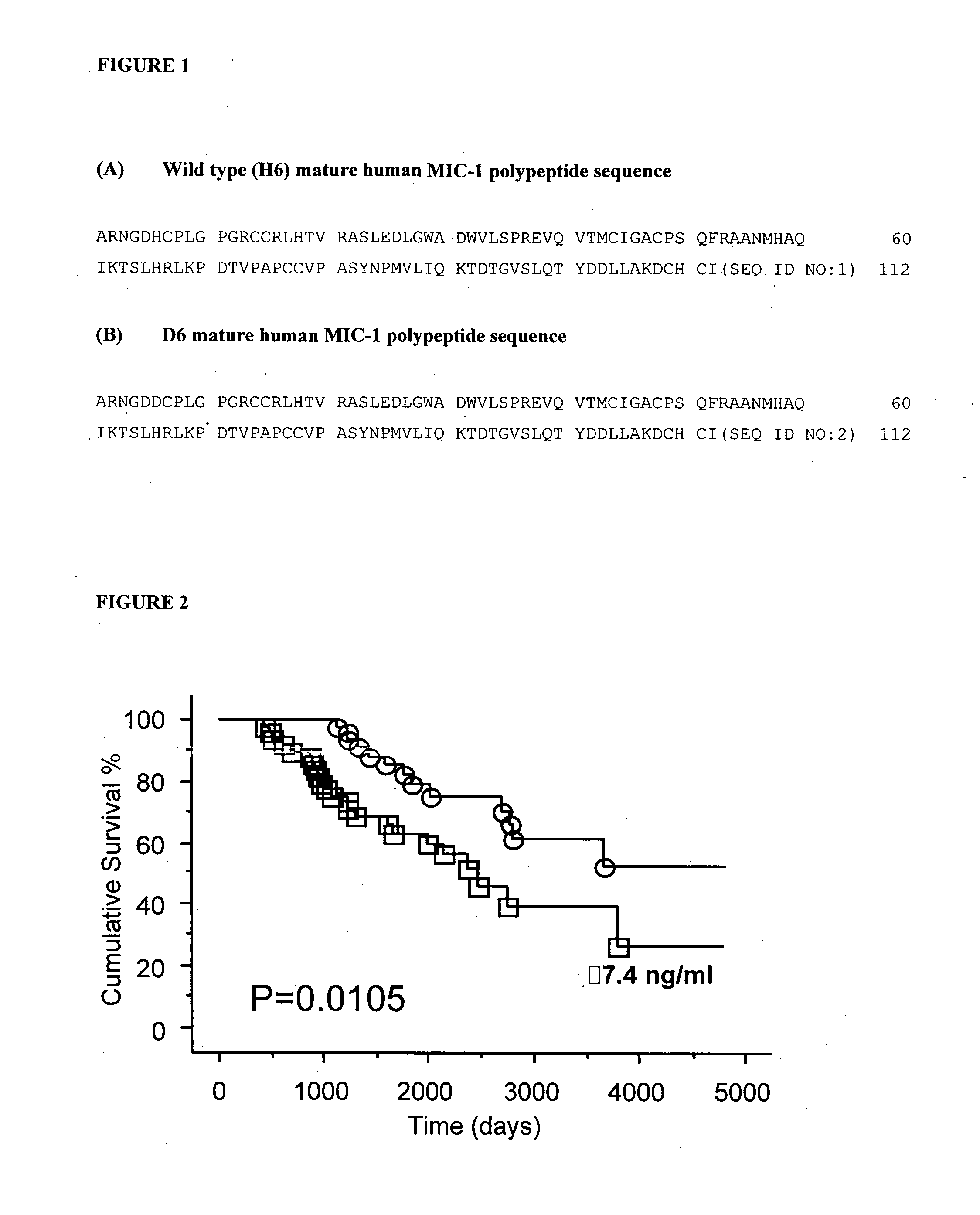
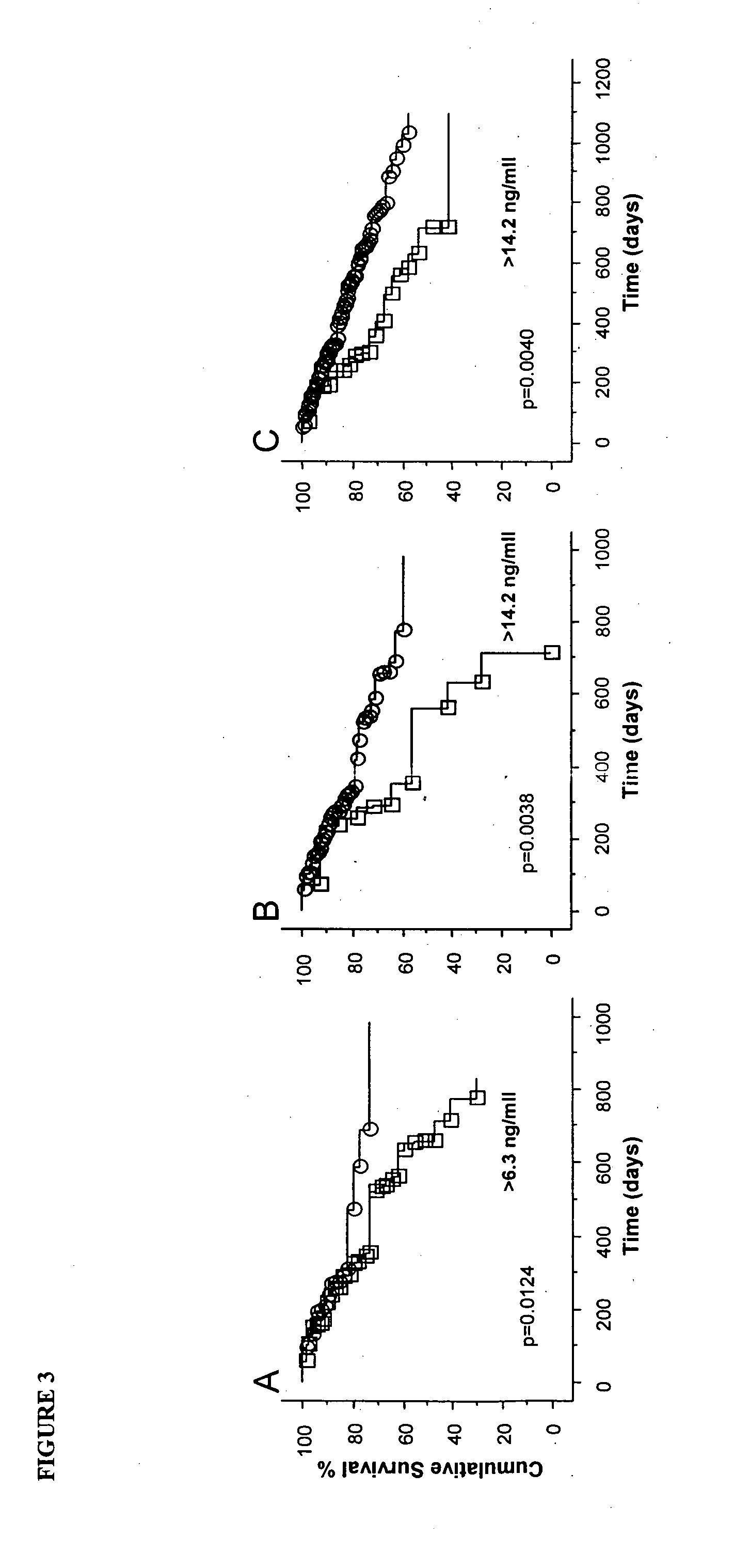
Methods of prognosis in chronic kidney disease
a kidney disease and prognosis technology, applied in the field of chronic kidney disease prognosis, can solve the problems of difficult to determine patients for whom a kidney transplant may be life-saving, many patients are likely to die, and the outcome and the nature of the link between them are ill-defined, so as to reduce the risk of death and prevent death
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Patient Cohorts
Materials and Methods
[0113]Cohorts of incident haemodialysis patients (ie patients that have reached the stage of requiring dialysis but have not yet commenced such treatment) from Sweden (n=98) and established haemodialysis patients from the United States of America (n=381) had serum MIC-1 and C-Reactive Protein (CRP)-4 levels and BMI measured at study entry. Serum CRP-4 (Dade Behring, Inc., Deerfield, Ill., United States of America) was determined as described in Brown et al. (2002a); and serum MIC-1 was determined as described below. Demographic information collected included age, sex, history of cardiovascular disease (CVD), history of diabetes mellitus (DM), height and weight. Additional markers of nutrition, body composition and inflammation were assessed in Swedish patients.
Swedish Cohort
[0114]Swedish patients were incident dialysis patients with end-stage renal disease enrolled in the dialysis program at the Karolinska University Hospital at Huddinge between 1...
example 2
Serum MIC-1 is Related to Subjective Global Nutritional Assessment (SGA) in the Swedish Cohort
[0121]Data regarding the US cohort has previously been published indicating that serum MIC-1 levels are negatively correlated with BMI (Johen et al., 2007). To further study the nutritional changes associated with elevated serum MIC-1 levels, the present applicant analysed data from Swedish patients, from whom nutritional information was available.
Results and Discussion
[0122]The SGA was segregated into normal (SGA=1, n=69) or abnormal (SGA>1, n=27) with data being unavailable for 2 patients. SGA was significantly related with S-creatinine (p=0.0050), BMI (p=0.0051), handgrip strength (p=0.0016 and serum MIC-1 level (p=0.0065). Patients with serum MIC-1 above the median (7430 pg / ml) were significantly more likely to have an abnormal SGA (p=0.0001, chi-square). Serum MIC-1 level above the median was independently associated with SGA in multivariate logistic regression (Table 3). This model wa...
example 3
Serum MIC-1 Level is Related to Nutrition and Markers of Inflammation and Oxidative Damage in the Swedish Cohort
[0124]The relationship between serum MIC-1 levels and circulating markers of inflammation (CRP, fibrinogen) and oxidation (8OHdG), muscle atrophy, BMI and age was examined.
Results and Discussion
[0125]As shown in Table 4, serum MIC-1 level was compared to the levels of circulating markers of inflammation (CRP, fibrinogen), oxidation (8-OHdG) and age in the Swedish population, as these factors may be indicators of mortality in end-stage renal disease. Initial univariate analysis using
[0126]Spearman rank correlation testing demonstrated that serum MIC-1 levels are related to age (ρ=0.252, p=0.0135), CRP (ρ=0.283, p=0.0056), fibrinogen (ρ=0.346, p=0.0018), 8-OHdG (ρ=0.308, p=0.0033) and serum albumin (ρ=−0.211, p=0.0388). Serum MIC-1 level and muscle atrophy did not significantly associate as a continuous variable; however, a strong trend was observed between serum MIC-1 above...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com