Diagnosis and treatment of multiple sulfatase deficiency and other sulfatase deficiencies
a technology of sulfatase and deficiency, applied in the direction of enzymology, drug composition, metabolic disorders, etc., can solve the problems of hampered efforts to identify this gene(s) and achieve the effect of increasing the c-formylglycine generating activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Multiple Sulfatase Deficiency is Caused by Mutations in the Gene Encoding the Human Cα-Formylglycine Generating Enzyme (FGE)
Experimental Procedures
Materials and Methods
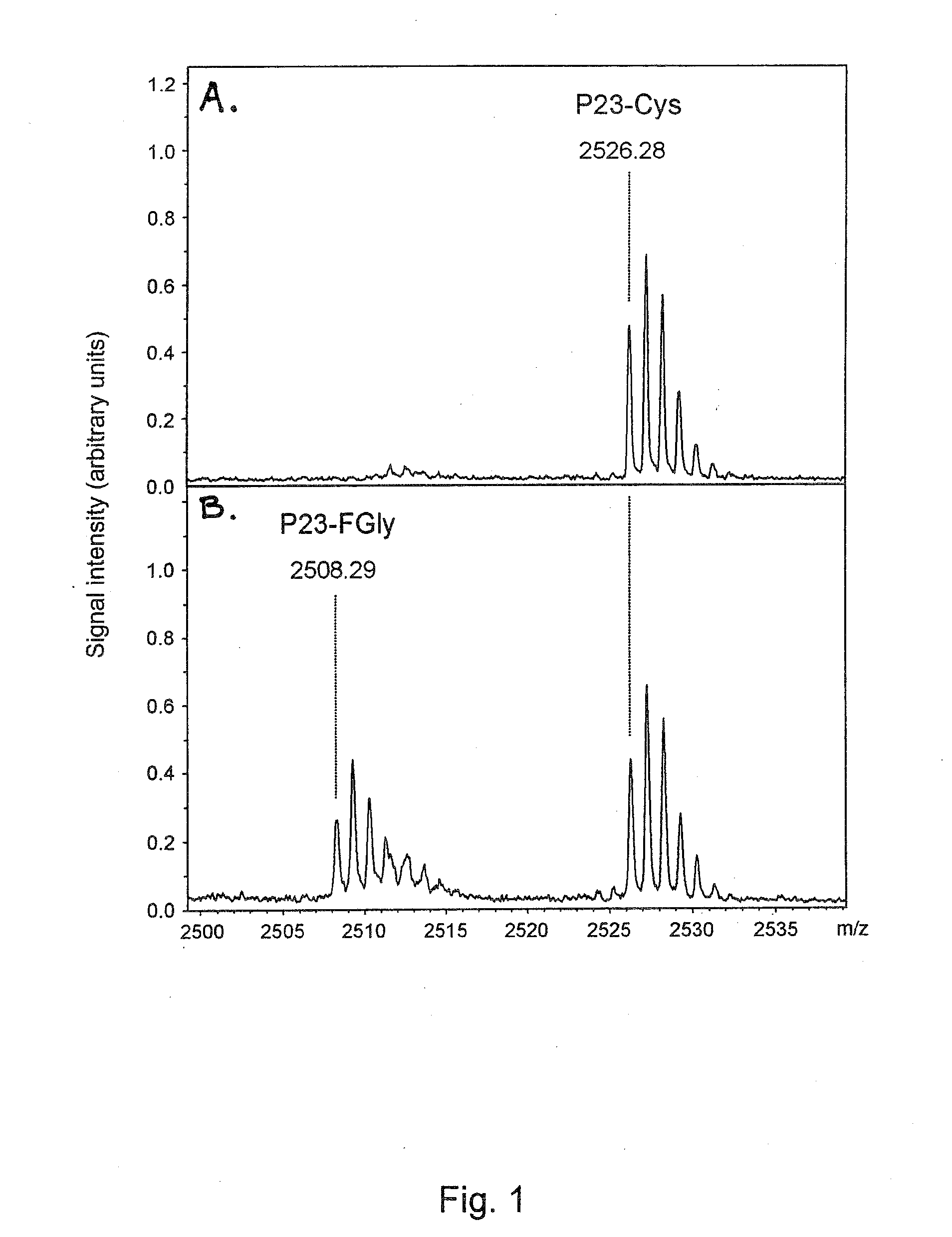
[0300]For monitoring the activity of FGE, the N-acetylated and C-amidated 23mer peptide P23 (MTDFYVPVSLCTPSRAALLTGRS) (SEQ ID NO:33) was used as substrate. The conversion of the Cysteine residue in position 11 to FGly was monitored by MALDI-TOF mass spectrometry. A 6 μM stock solution of P23 in 30% acetonitrile and 0.1% trifluoroacetic acid (TFA) was prepared. Under standard conditions 6 pmol of P23 were incubated at 37° C. with up to 10 μl enzyme in a final volume of 30 μl 50 mM Tris / HCl, pH 9.0, containing 67 mM NaCl, 15 μM CaCl2, 2 mM DTT, and 0.33 mg / ml bovine serum albumin. To stop the enzyme reaction 1.5 μl 10% TFA were added. P23 then was bound to ZipTip C18 (Millipore), washed with 0.1% TFA and eluted in 3 μl 50% acetonitrile, 0.1% TFA. 0.5 μl of the eluate was mixed with 0.5 μl of matri...
example 2
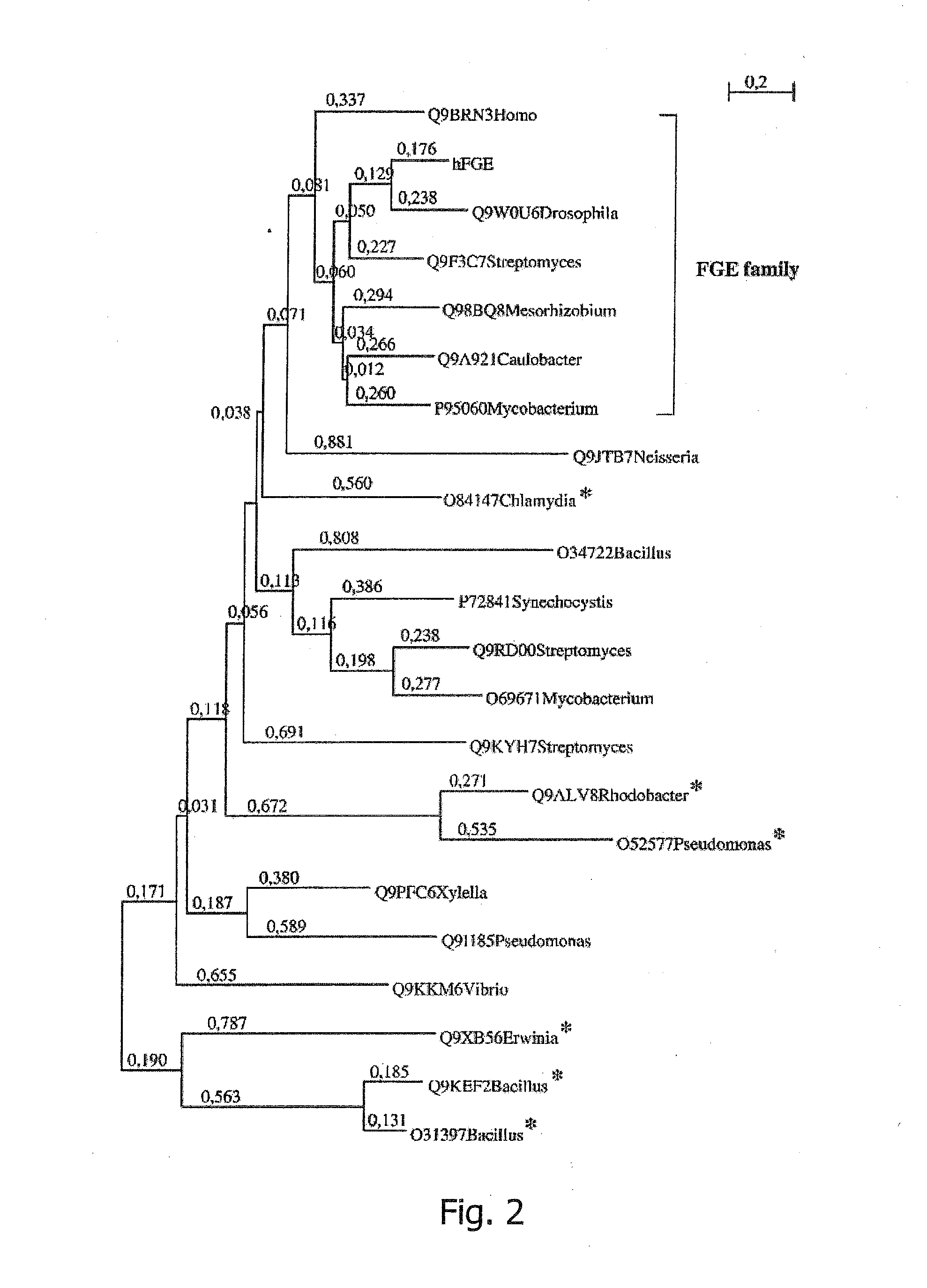
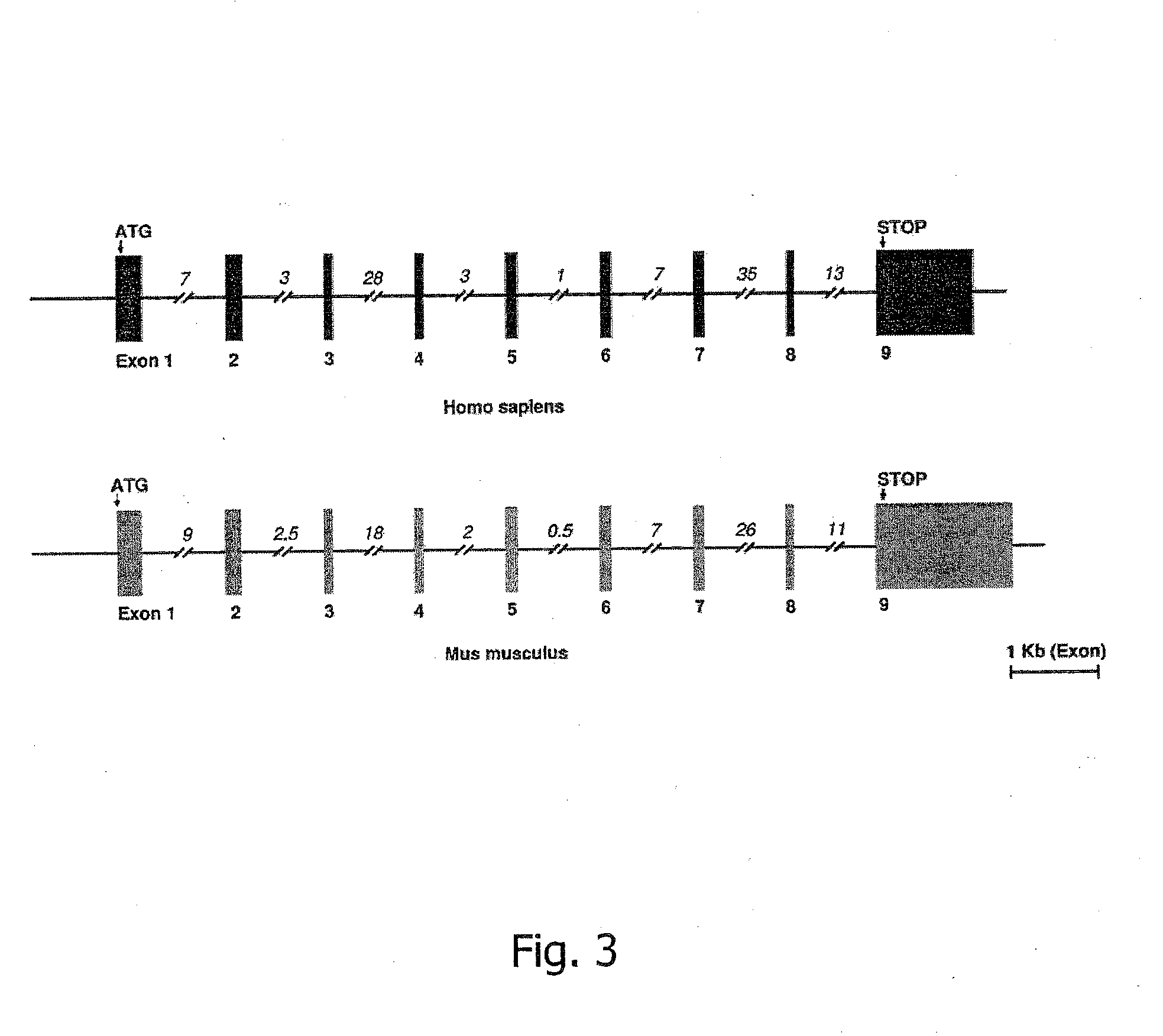
[0324]The Human FGE Gene Defines a New Gene Family Modifying Sulfatases which is Conserved from Prokaryotes to Eukaryotes
[0325]Signal peptides and cleavage sites were described with the method of von Heijne (Nucleic Acids Res., 1986, 14:4683) implemented in EMBOSS (Rice et al., Trends in Genetics, 2000, 16:276-277), and the method of Nielsen et al. (Protein Engineering, 1997, 10:1-6). N-glycosylation sites were predicted using the algorithm of Brunak (Gupta and Brunak, Pac. Symp. Biocomput., 2002, 310-22).
[0326]Functional domains were detected by searching PFAM-Hidden-Markov-Models (version 7.8) (Sonnhammer et al., Nucleic Acids Res., 1998, 26:320-322). Sequences from the PFAM DUF323 seed were obtained from TrEMBL (Bairoch, A. and Apweiler, R., Nucleic Acids Res., 2000, 28:45-48). Multiple alignments and phylogenetic tree constructions were performed with Clustal W (Thompson, J., et al., Nucleic Acids Res., 1994, 22:4673-4680). For phylogenetic tree computation, gap po...
example 3
[0348]FGE Expression Causes Significant Increases in Sulfatase Activity in Cell Lines that Overexpress a Sulfatase
[0349]We wanted to examine the effects of FGE on cells expressing / overexpressing a sulfatase. To this end, HT-1080 cells expressing human sulfatases Iduronate 2-Sulfatase (I2S) or N-Acetylgalactosamine 6-Sulfatase (GALNS) were transfected in duplicate with either a FGE expression construct, pXMG.1.3 (Table 7 and FIG. 4) or a control plasmid, pXMG.1.2 (FGE in antisense orientation incapable of producing functional FGE, Table 7). Media samples were harvested 24, 48, and 72 hours following a 24 hour post-electroporation medium change. The samples of medium were tested for respective sulfatase activity by activity assay and total sulfatase protein level estimated by ELISA specific for either Iduronate 2-Sulfatase or N-Acetylgalactosamine 6-Sulfatase.
TABLE 6Transfected Cell Lines Expressing Sulfatases Used as Substrates forTransfectionCell StrainPlasmidSulfatase Expressed36Fp...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com