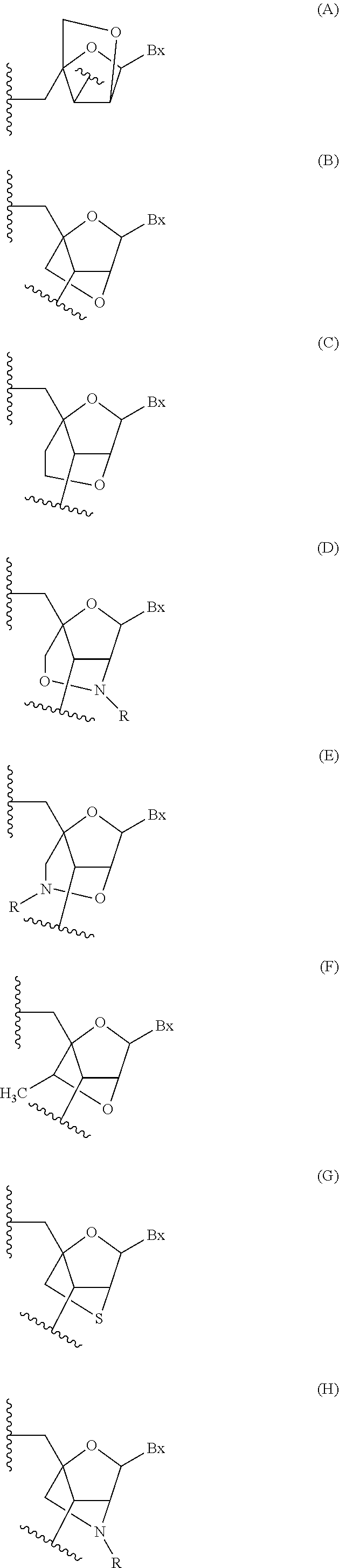
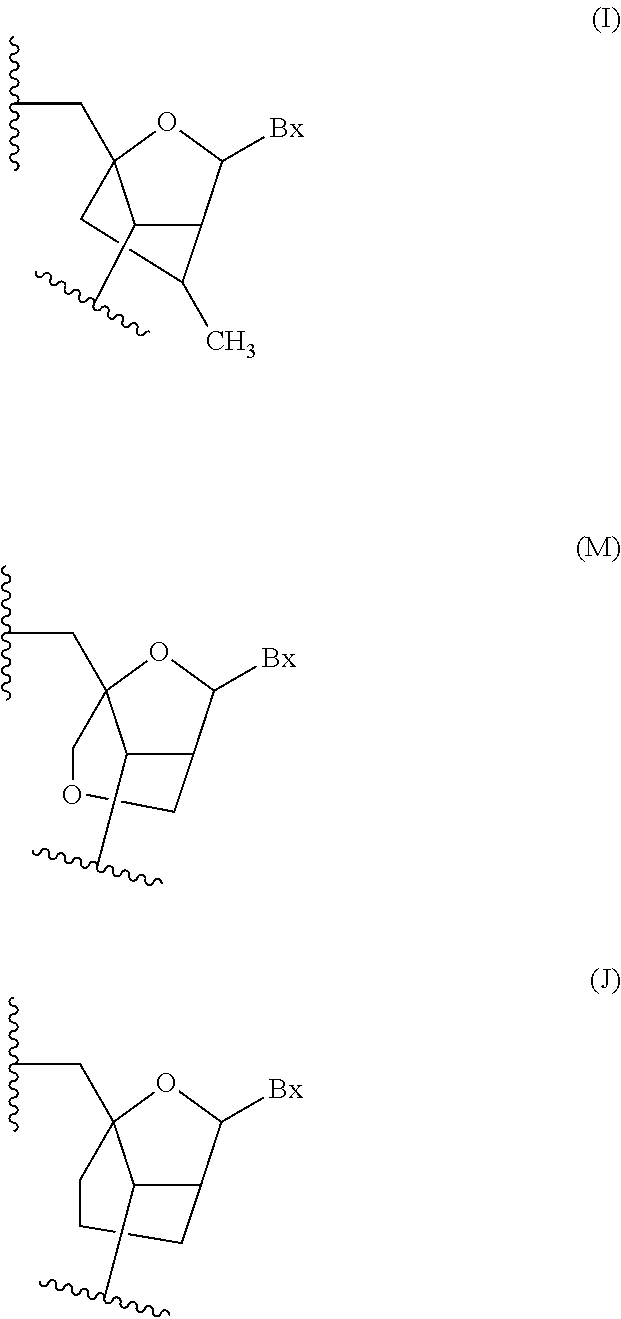
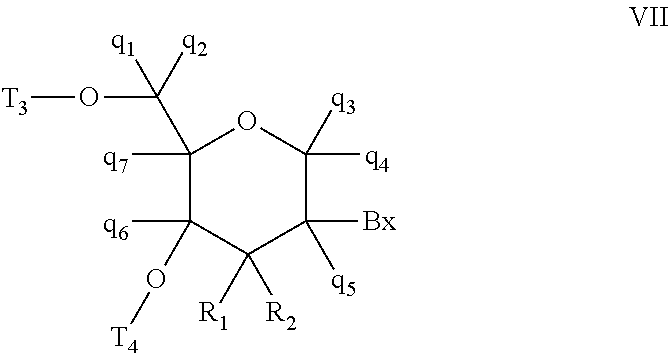
Oligomeric compounds comprising bicyclic nucleosides and uses thereof
a technology of bicyclic nucleosides and compounds, applied in the field ofoligomeric compounds comprising bicyclic nucleosides, can solve the problems of cleavage of mrna and interfere with polyadenylation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Antisense Inhibition of Human Target X in HuVEC Cells
[0349]Antisense oligonucleotides were designed to target a messenger RNA molecule (Target X) and were tested for their effects on target X mRNA in vitro. Cultured HuVEC cells at a density of 20,000 cells per well were transfected using electroporation with 250 nM antisense oligonucleotide. After a treatment period of approximately 24 hours, RNA was isolated from the cells and Target X mRNA levels were measured by quantitative real-time PCR. Target X mRNA levels were adjusted according to total RNA content, as measured by RIBOGREEN®. Results are presented as percent inhibition of Target X, relative to untreated control cells. A total of 40 oligonucleotides were tested. Only those oligonucleotides which were selected for further study are shown in Table 4.
[0350]The ‘Chemistry’ column describes the sugar modifications of each oligonucleotide. ‘k’ indicates an (S)-cEt sugar modification; the number indicates the number of deoxynucleo...
example 2
Dose-Dependent Antisense Inhibition of Human Target X in HuVEC Cells
[0351]Gapmers from the studies described above exhibiting significant in vitro inhibition of Target X mRNA were selected and tested at various doses in HuVEC cells. Cells were plated at a density of 20,000 cells per well and transfected using electroporation with 31.3 μM, 62.5 μM, 125.0 μM, 250.0 μM, 500.0 μM, and 1000.0 μM concentrations of antisense oligonucleotide, as specified in Table 5. After a treatment period of approximately 16 hours, RNA was isolated from the cells and Target X mRNA levels were measured by quantitative real-time PCR. Target X mRNA levels were adjusted according to total RNA content, as measured by RIBOGREEN®. Results are presented as percent inhibition of Target X, relative to untreated control cells. The antisense oligonucleotides were tested in a series of experiments that had similar culture conditions. The results for each experiment are presented in separate tables shown below.
[0352]T...
example 3
Dose-Dependent Antisense Inhibition of Human Target X in HuVEC Cells
[0353]Additional antisense oligonucleotides were designed as deoxy, MOE and (S)-cEt oligonucleotides targeting Target X gene sequences and were tested at various doses in HuVEC cells. The ‘Chemistry’ column describes the sugar modifications of each oligonucleotide. ‘k’ indicates an (S)-cEt sugar modification; the number indicates the number of deoxynucleosides; otherwise indicates deoxyribose; and ‘e’ indicates a MOE modification. The internucleoside linkages throughout each gapmer are phosphorothioate linkages. All cytosine residues throughout each gapmer are 5-methylcytosines. “Start site” indicates the 5′-most nucleoside to which the gapmer is targeted in the human gene sequence. “Stop site” indicates the 3′-most nucleoside to which the gapmer is targeted human gene sequence. Each gapmer listed in Table 6 is targeted to the human Target X genomic sequence. Oligonucleotides having the same start site and stop site...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pharmaceutical composition | aaaaa | aaaaa |
| chemical analysis | aaaaa | aaaaa |
| chemical | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com