Diagnostic for Sepsis
a sepsis and sepsis technology, applied in the field of diagnosis, can solve the problems of poorly characterized relations, achieve the effect of decreasing reducing the risk of organ failure, and reducing the risk of a subj
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Definition and Characterization of the Signature
[0231]Confirmed Sepsis Patients Express an “Endotoxin Tolerance Signature.”
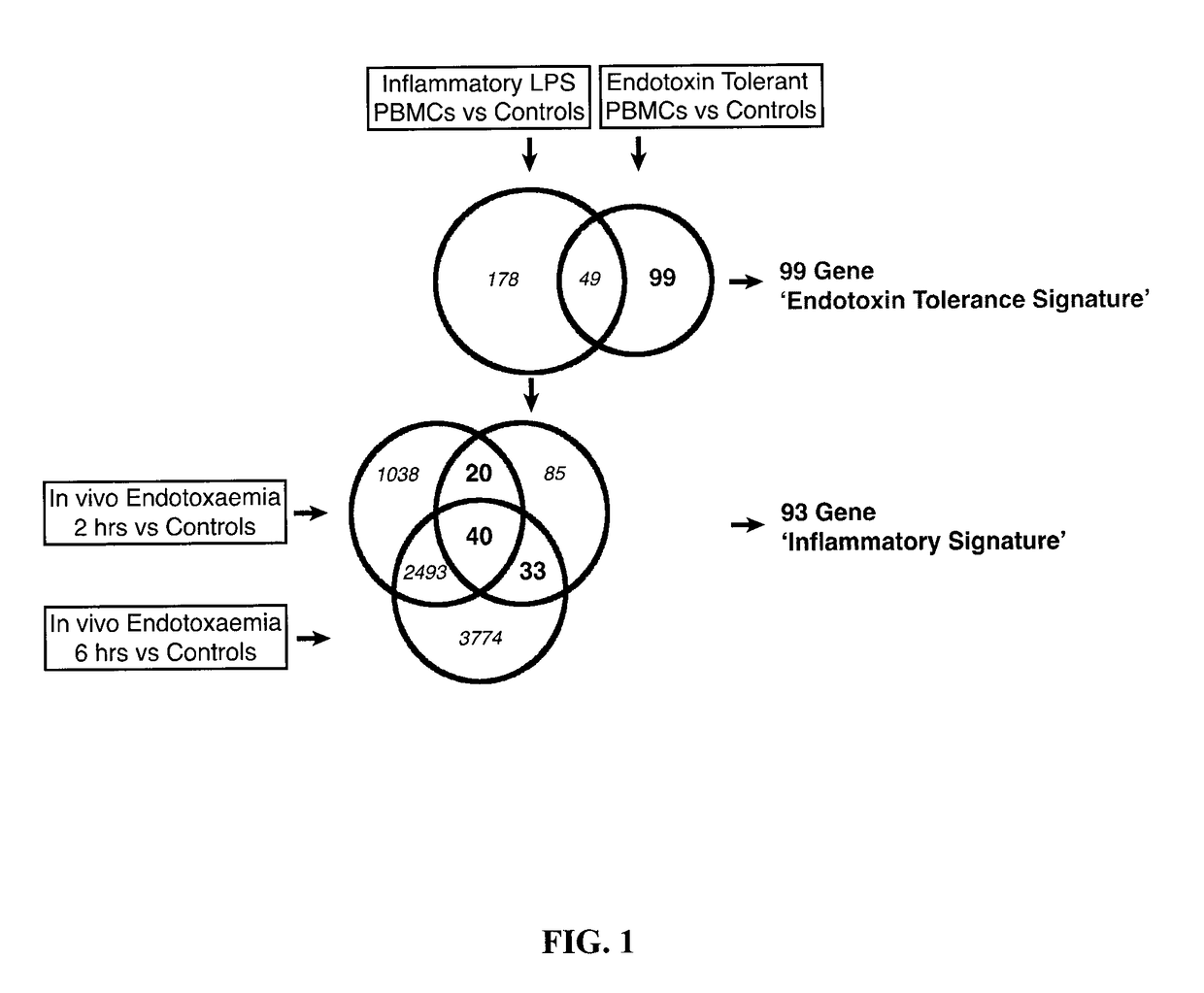
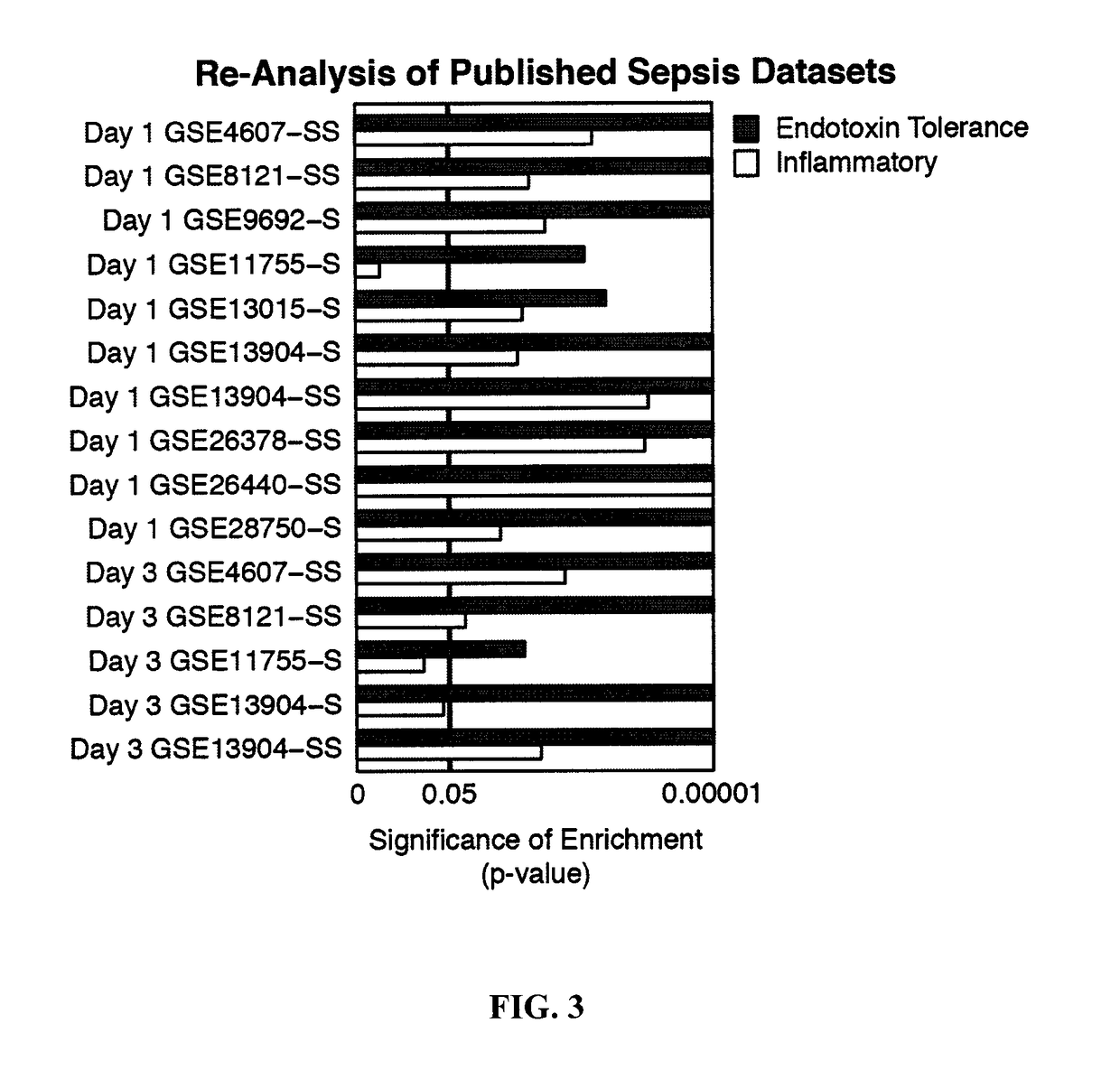
[0232]To characterize the development of the immunosuppressive stage in sepsis and to conclusively determine its links with endotoxin tolerance, a robust bioinformatics approach was taken. To define an endotoxin tolerance gene signature, microarray analyses of human peripheral blood mononuclear cells (PBMC) treated either once with LPS to model inflammatory signalling, or twice to model endotoxin tolerance was used. An “Endotoxin Tolerance Signature” (Table A below), comprising 99 genes was identified based on genes uniquely differentially expressed in endotoxin-tolerant PBMCs, but not inflammatory PBMCs, as compared to controls. For comparison, we defined an “Inflammatory Signature” from previous PBMC microarray data (Pena et al., 2011) and an in vivo experimental endotoxemia dataset (Calvano et al., 2005) (FIG. 1, Table 4). Having defined a genetic signature f...
example 2
New Therapies Based on the Endotoxin Tolerance Signature
[0252]Network analysis of the Endotoxin Tolerance genes revealed that most of the genes formed a very tight subnetwork strongly suggesting that the signature reflects critical mechanisms likely related to immune dysfunction in sepsis patients (FIG. 8).
[0253]One implication of knowing that a patient is going to soon suffer from sepsis is that one can apply an appropriate antibiotic therapy comprising a cocktail of the most potent drugs. Current clinical guidelines indicate that while waiting for culture results, a patient should be started on intravenous ceftriaxone and azithromycin. The purpose of this regimen is to try to avoid major resistance issues since only a portion of the patients who are thought to have the potential to acquire sepsis actually do so (see e.g. Table 3). Knowing that a patient has sepsis very early in the course of disease would enable physicians to prescribe the most aggressive therapies to try to reduc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| nucleic acid | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com