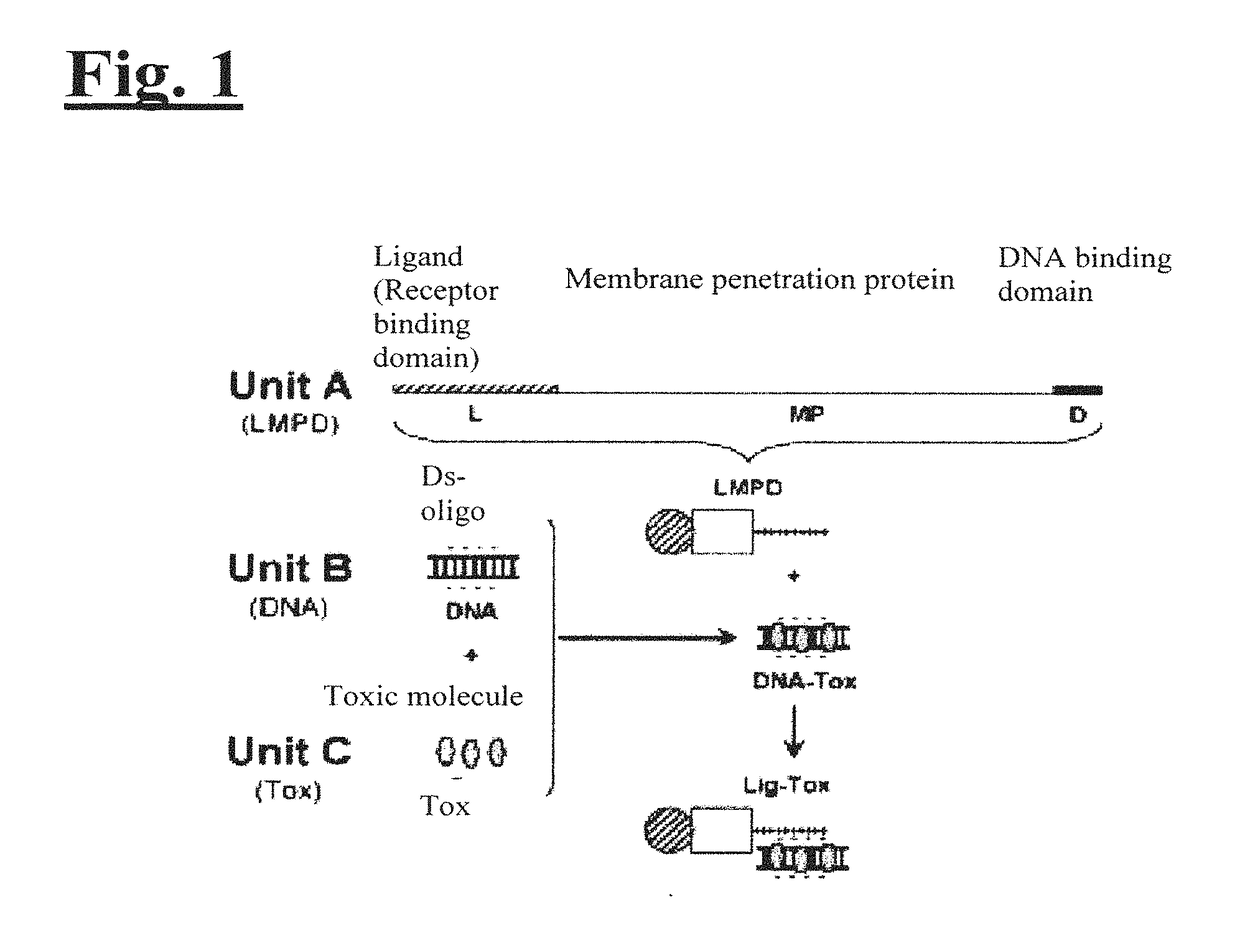
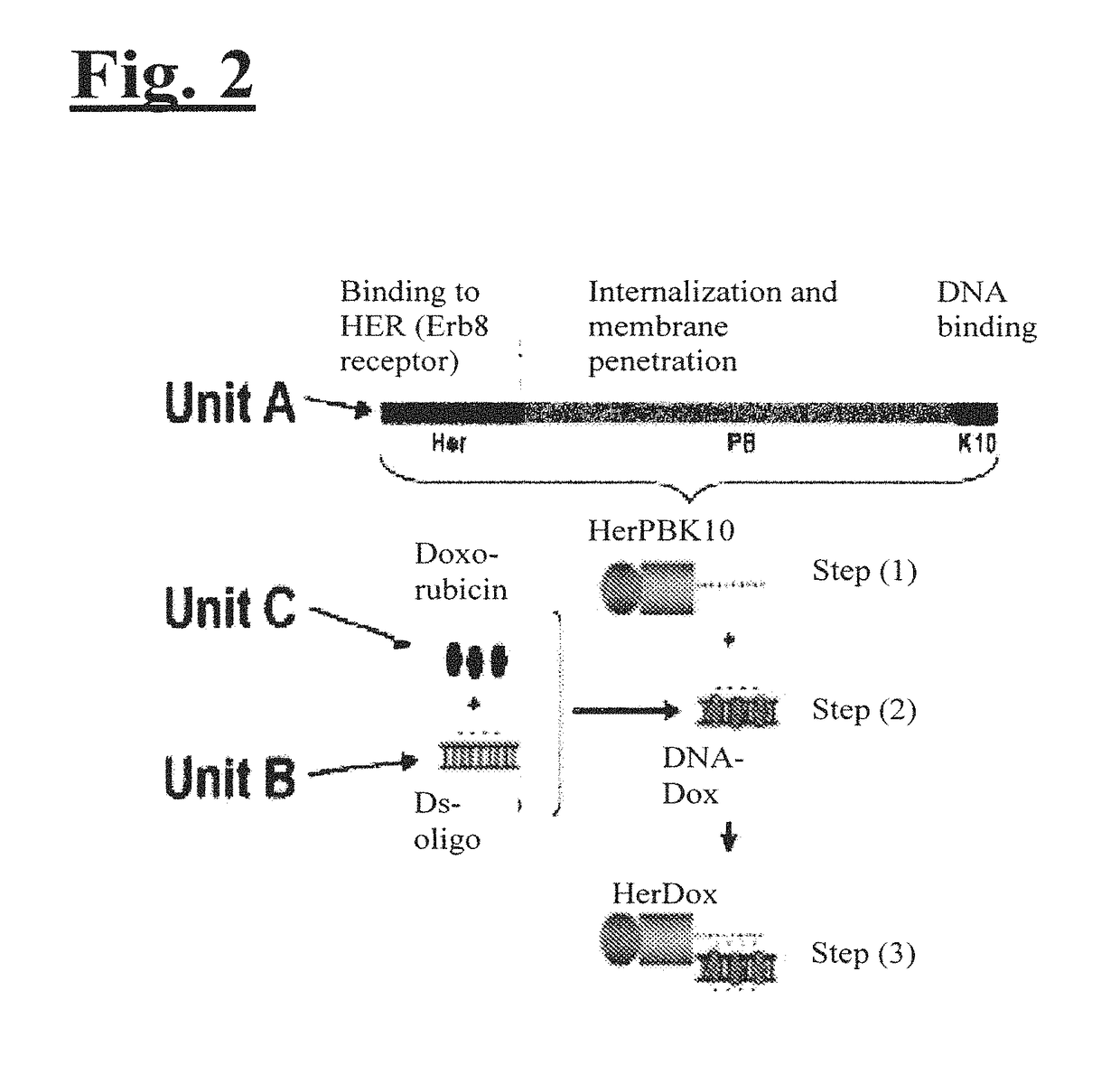
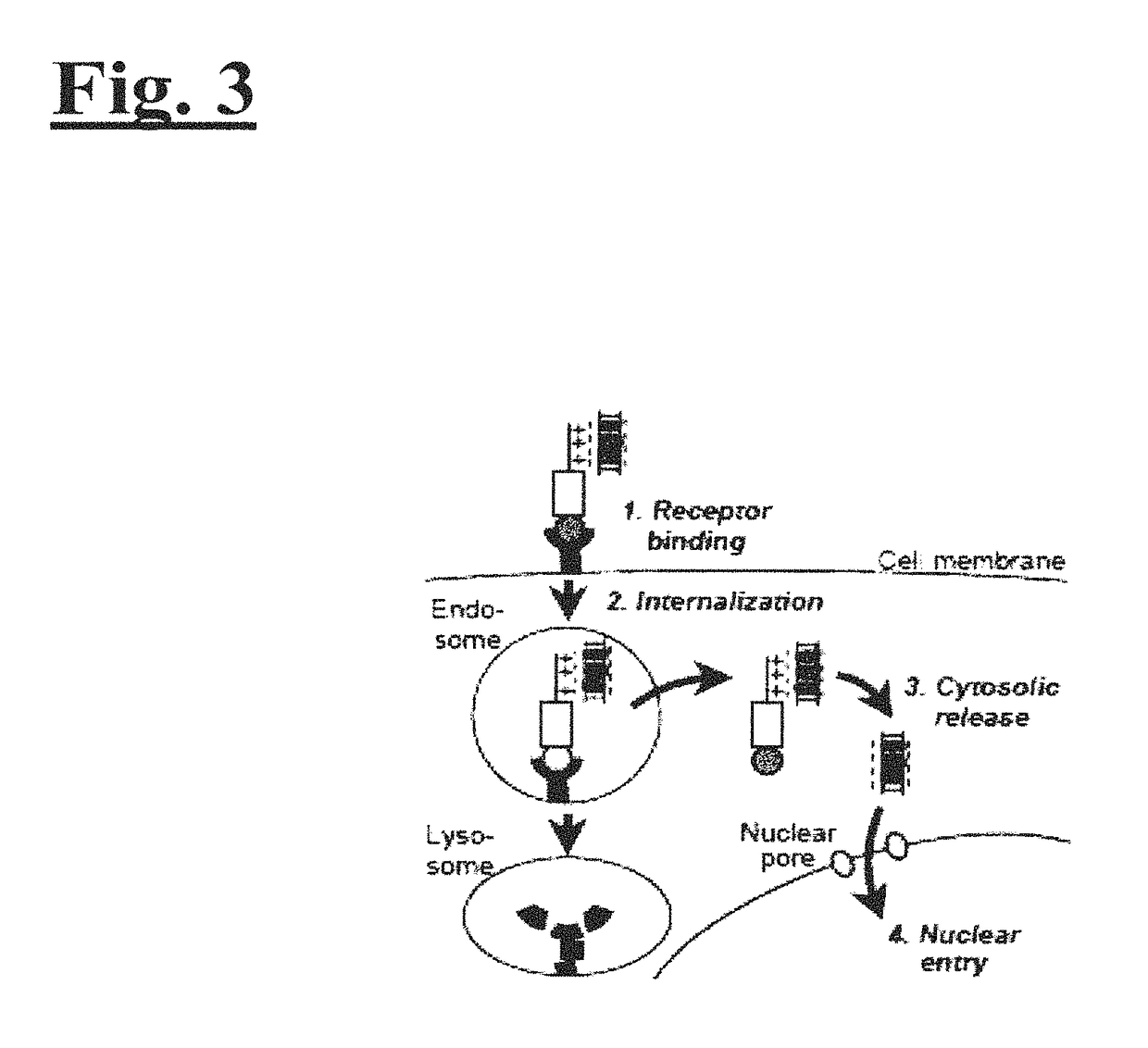
Targeting trastuzumab-resistant her2+ breast cancer with a her3-targeting nanoparticle
a technology of nanoparticles and breast cancer, applied in the field of therapeutics, can solve the problems of poor patient survival, resistance of a large portion of patients, and very few treatment options
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Targeted Delivery of Chemotherapeutic to HER2+ Breast Cancer Cells
[0204]The disclosed technology was tested on HER2+ breast cancer cells in vitro and in vivo. As illustrated in FIG. 2, to engineer the drug delivery molecule to target HER2+ breast cancer, Unit A includes a protein called HerPBK10, which is generated by the methods described in L. K. Medina-Kauwe et al. Gene Therapy, 8:1753-1761 (2001), incorporated by reference herein in its entirety. HerPBK10 contains the receptor binding domain of heregulin fused to the cell penetrating adenovirus penton base protein modified by a carboxy (C)-terminal decalysine. The ‘Her’ segment of HerPBK10 is obtained from the receptor binding domain of heregulin-α, which binds specifically to HER2 / HER3 or HER2 / HER4 subunit heterodimers. Although heregulin interacts directly with HER3 or HER4, but not HER2, ligand affinity is greatly enhanced by HER2. Thus, tumor cells that over-express HER2 (i.e., HER2+ tumor cells) are believed to be good cand...
example 2
HerDox is Highly Stable During Assembly
[0208]HerDox consists of three components: Dox; a small double-stranded nucleic acid (which is directly responsible for carrying Dox); and the targeted protein, HerPBK10. HerDox is assembled in two steps. First, Dox is mixed with the DNA to form a DNA-Dox pair by DNA intercalation. Then, the DNA-Dox pair is mixed with HerPBK10 to form HerDox by electrophilic interaction. To separate DNA-Dox from free Dox, the mixture underwent ultrafiltration centrifugation. The inventors found that >95% of the Dox added to the DNA did not release from the DNA during the ultrafiltration spin, indicating high retention of the drug even during a high speed spin (FIG. 4(a)). The absorbance spectra of retentate and filtrate from this spin confirm that the absorbance maximum of retentate coincides with unfiltered Dox, whereas no such absorbance is detectable in the filtrate (FIG. 4(b)). The retentate was then incubated with HerPBK10 and the resulting HerDox complex ...
example 3
HerDox is Highly Stable During Storage and in Serum
[0209]The inventors tested the stability of HerDox over 12 days under different storage temperatures: 40° C., room temperature, or 37° C. On each day, a sample underwent ultrafiltration, then filtrates and retentates were measured to determine whether any Dox was released from the complex. At 4° C., 100% of the product remained intact up to 12 days, and, interestingly, room temperature and 37° C. appeared to enhance the incorporation of the drug into the HerDox product (FIGS. 6(a) and 6(b)). Altogether, these findings suggest that HerDox remains stable and does not release Dox after prolonged storage under different temperatures. The inventors also examined HerDox stability in serum-containing media at 37° C. HerDox immobilized on nickel sepharose (via the HerPBK10 histidine tag) was incubated at 37° C. in complete (i.e. 10% fetal bovine serum-containing) media (to mimic tissue culture conditions) for different time periods before t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com