Methods and compositions for transposition using minimal segments of the eukaryotic transformation vector piggybac
a technology of eukaryotic transformation vector and eukaryotic transformation vector, applied in the field of transposon piggybac, can solve the problems of limited gene transfer length, complicated procedure, and limited effective size of genes that may be inserted, and achieve the effect of enhancing the infusion of dna molecules into cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
istance Required Between Termini for Movement of a PiggyBac Transposon Construct
[0109]The interplasmid transposition assay was carried out essentially as previously described by Lobo et al. (1999), Thibault et al. (1999) and Sarkar et al. (1997a). Embryos were injected with a combination of 3 plasmids. The donor plasmid, pB(KOα), carried a piggyBac element marked with the kanamycin resistance gene, ColE1 origin of replication, and the lacZ gene. The transposase providing helper plasmid, pCaSpeR-pB-orf, expressed the full length of the piggyBac ORF under the control of the D. melanogaster hsp70 promoter. The target B. subtilis plasmid, pGDV1, is incapable of replication in E. coli, and contains the chloramphenicol resistance gene. Upon transposition of the genetically tagged piggyBac element from pB(KOα) into the target plasmid pGDV1 with the help of the transposase provided by the helper pCaSpeR-pB-orf that expresses the piggyBac transposase protein from a minimal hsp70 promoter (se...
example 3
mid Transposition Assay of pCRII-ITR and pBSII-ITR Plasmids
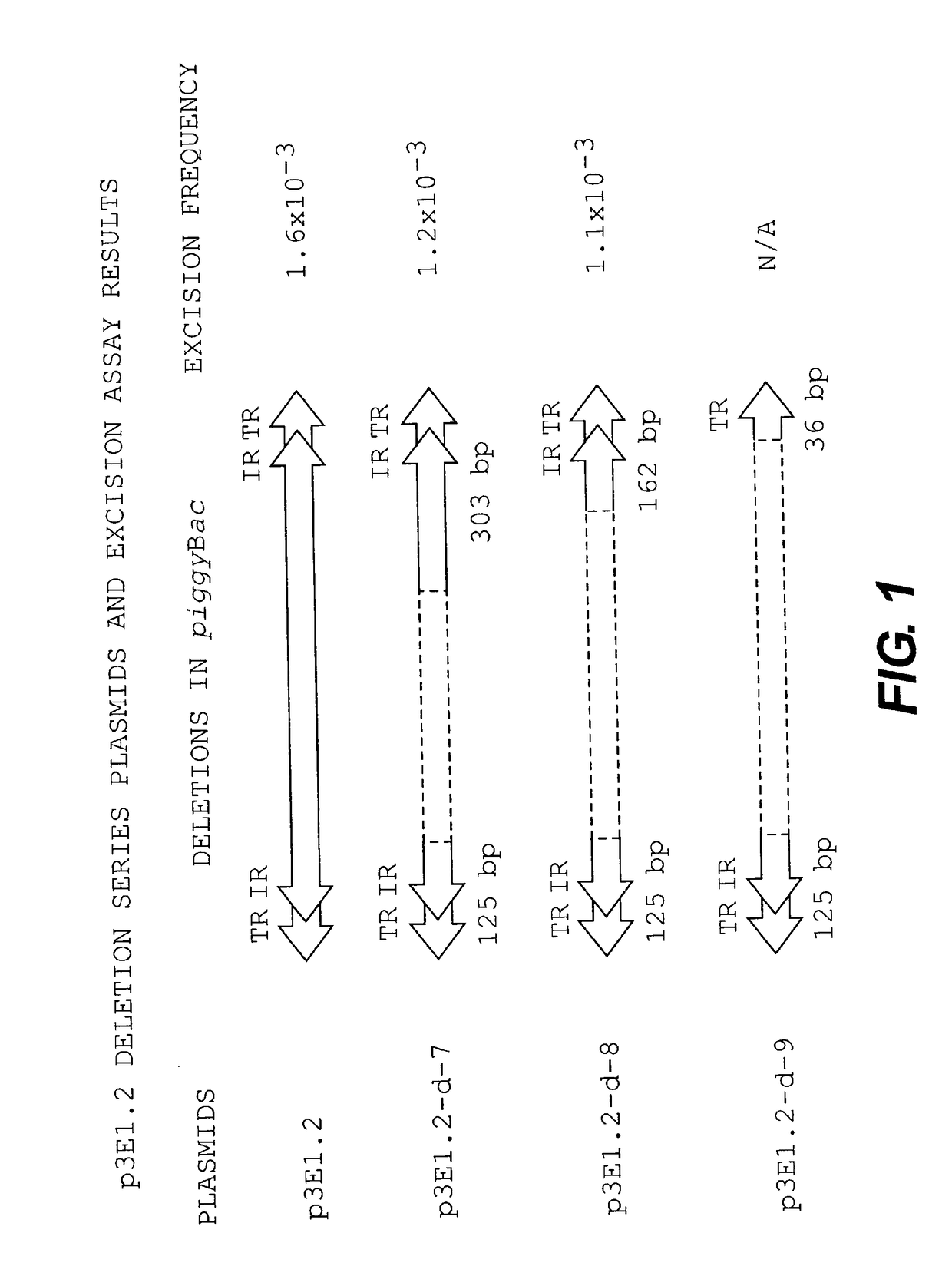
[0112]According to an embodiment of the present invention, the excision assay described herein shows that a minimum of 163 bp of the 3′ terminal region and 125 bp of the 5′ terminal region (from the restriction site SacI to the end of the element) may be used for excision, while the pIAO-P / L constructs showed that a minimal distance of 55 bp between termini may be utilized to effect movement. These data suggested that the inclusion of intact left and right terminal and internal repeats and spacer domains would be sufficient for transposition.
[0113]The pCRII-ITR plasmid was constructed following PCR of the terminal domains from pIAO-P / L-589 using a single IR specific primer. A second construct pCRII-JFO3 / 04 was also prepared using two primers that annealed to the piggyBac 5′ and 3′ internal domains respectively, in case repeat proximate sequences were required.
[0114]The interplasmid transposition assay was performed in T. ni ...
example 4
ion of Minimum PiggyBac Vector pXL-Bac
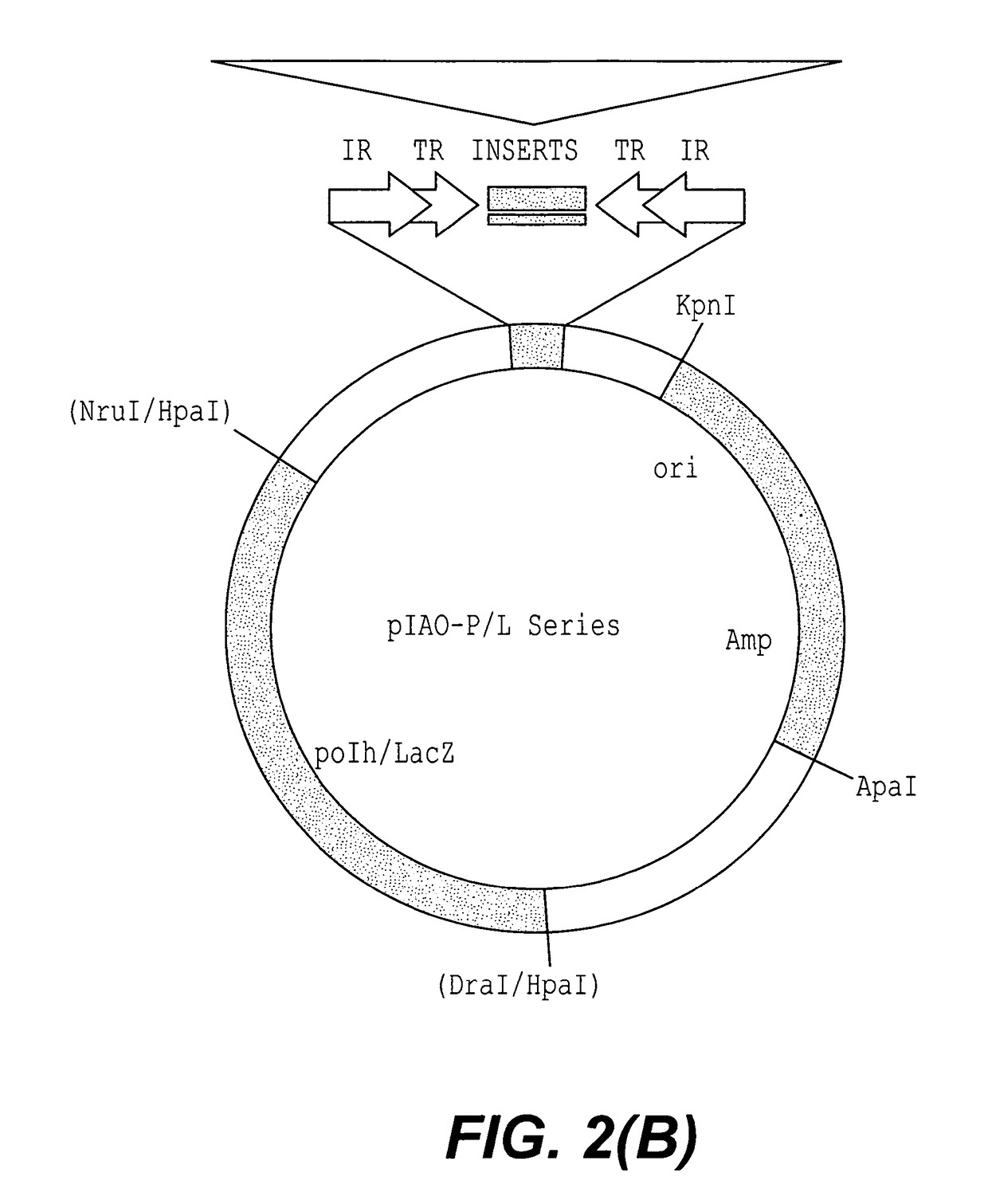
[0116]A new piggyBac minimum vector pXL-Bac (FIG. 3(C2)) was also constructed by combining the 702 bp BamHI ITR fragment with the pBlueScript II BamHI fragment and inserting a PCR amplified pBSII multiple cloning site (MCS) between the terminal repeats. The pXL-Bac vector was tested by inserting an XbaI fragment from πKOα (obtained from A, Sarkar, University of Notre Dame), containing the Kanamycin resistance gene, E. coli replication origin, and Lac a-peptide, into the MCS of pXL-Bac to form pXL-Bac-KOa. Interplasmid transposition assays yielded a frequency of over 10−4 for transposition of the modified ITR sequence, a similar level as observed for the intact piggyBac element.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com