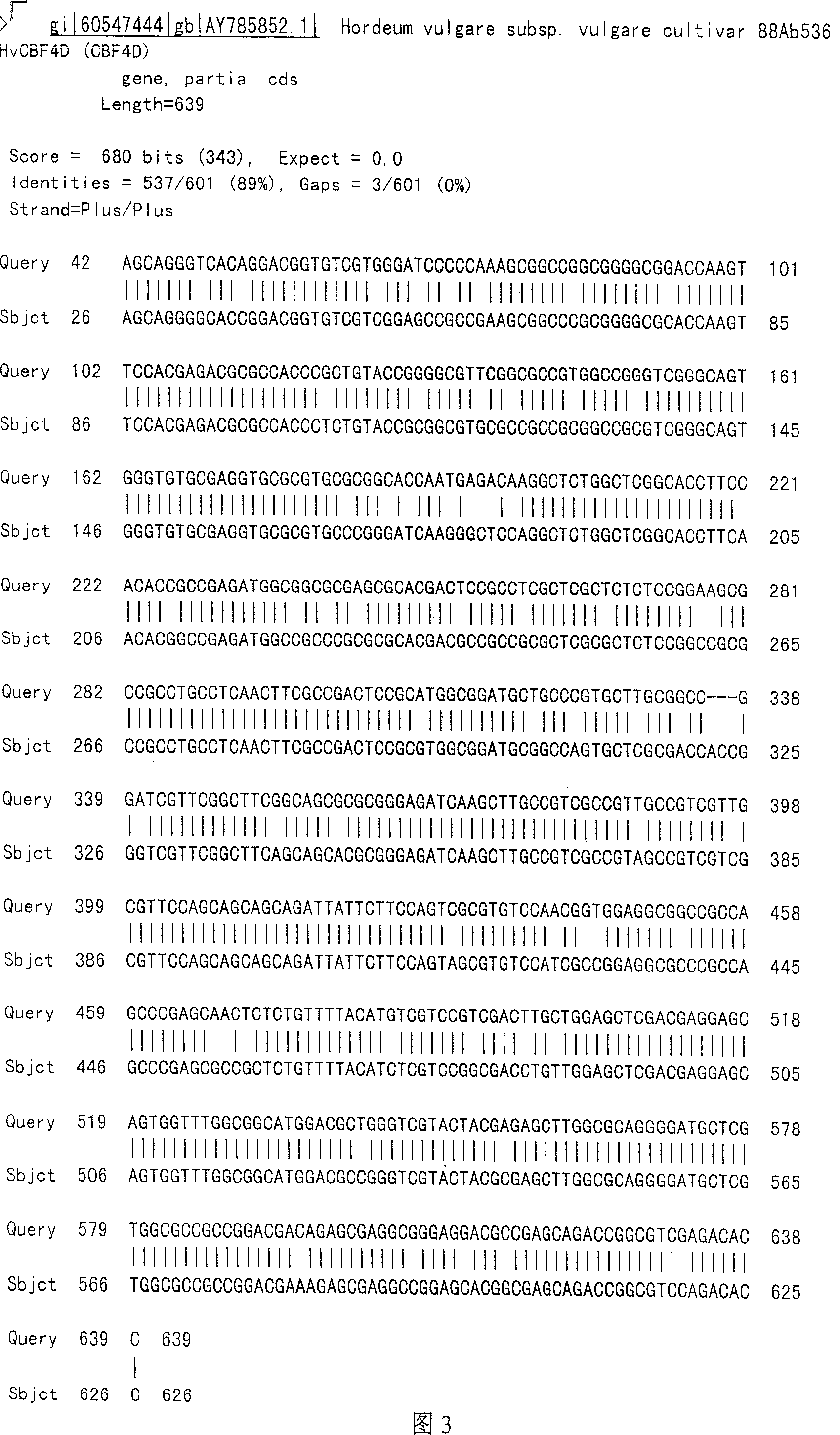
cDNA sequence of dehydrolysis responding transcription factor DREB gene of wheal
A technology of transcription factors and genes, applied in the field of agricultural biological genetic engineering, can solve problems such as difficulty in improving plant stress resistance and difficulty in researching the stress resistance of cultivated wheat
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] (1) Plant material and treatment:
[0037] The seeds of cultivated wheat Jing 411 were sterilized with 10% antifumin, soaked in deionized water, and then cultured at 23°C in the dark for 6-10 days. The leaves were harvested for DNA extraction. The wheat seedlings of cold-treated samples were subjected to cold stress at 4°C for 2 hours before RNA extraction.
[0038] (2) Extraction of Wheat Jing 411 Genomic DNA
[0039]Grind 0.2g of etiolated seedlings into powder in liquid nitrogen; add 2ml of DNA extraction buffer (500mM NaCl, 100mM Tris HCl, 50mM EDTA), bathe in 65°C water for 15 minutes, ice bath for 10 minutes, centrifuge at 3,000rpm 4°C for 15 minutes , remove tissue pieces; take the supernatant, add an equal volume of phenol and centrifuge at 3,000rpm 4°C for 10 minutes; take the supernatant, add an equal volume of phenol and centrifuge at 3,000rpm 4°C for 10 minutes; Centrifuge for 10 minutes; take the supernatant, add 2 times the volume of cold absolute ethano...
Embodiment 2
[0041] (1) Wheat Jing 411 DREB gene fragment (WtJ 292 ) PCR amplification
[0042] 1. WtJ 292 Fragment PCR amplification
[0043] 1) Design of primers
[0044] Three DREB gene sequences (gi|17148646, gi|17148648, gi|17148650) of published rye were queried on GenBank, and multiple sequences were compared, and the primer design software Primer 5.0 was used to find and synthesize ( All the following primers were entrusted to Shanghai Sangong to synthesize) The primers are as follows:
[0045] Forward1: 5'TGC CTC AAC TTC GCC GAC TCC3' (SEQ ID NO.3)
[0046] Reverse1: 5'CGA GCA TCC CCT GCG CCA AG3' (SEQ ID NO.4)
[0047] 2) PCR amplification reaction system: 10×Buffer 2.5μl, MgCl 2 (25mM) 1.5μl, dNTP (10mM) 0.5μl, Forward1 (10μM) 0.25μl, Reverse1 (10μM) 0.25μl, DNA Template 100ng, Taq (5U / μl) 0.25μl, ddH 2 O make up to 25 μl. PCR parameters: pre-denaturation at 95°C for 2 minutes; denaturation at 95°C for 1 minute, annealing at 60°C for 30 seconds, extension at 72°C for 30 ...
Embodiment 3
[0078] Embodiment 3: the extraction of whole wheat RNA
[0079] The total RNA of wheat was extracted by acidic guanidine isothiocyanate method: Weigh 0.2 g of yellowed wheat seedlings, wash them with sterilized double distilled water, dry them with sterilized filter paper, cut them into pieces in a pre-cooled mortar; add liquid nitrogen and grind until In powder form, add 2ml of Solution D, grind with the powder until clear; (the following steps are operated on ice) pour into a 7ml centrifuge tube, add 200μl 2mol / L NaAC (PH4.0) and invert several times to mix well and place on ice ; Add an equal volume of water-saturated phenol, shake and mix vigorously; add 400 μl chloroform-isoamyl alcohol (49:1), shake and mix vigorously; bathe in ice for 30 minutes, invert several times during the period, it is advisable to avoid stratification; Centrifuge at 11,000rpm at 4°C for 15 minutes; take the supernatant, add an equal volume of phenolform, extract again, and centrifuge at 11,000rpm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com