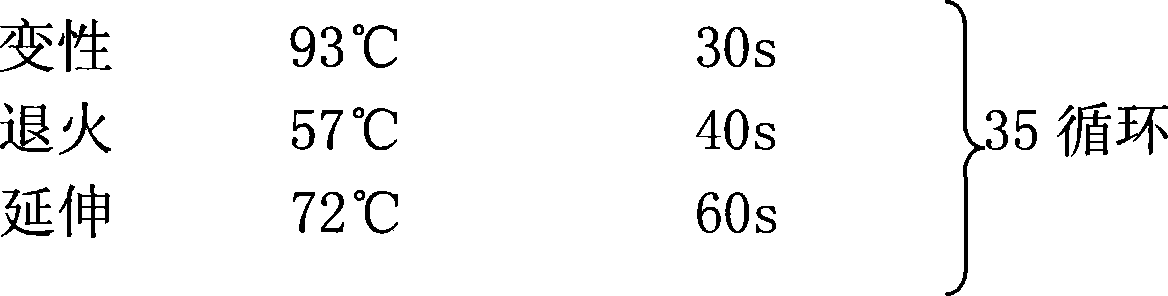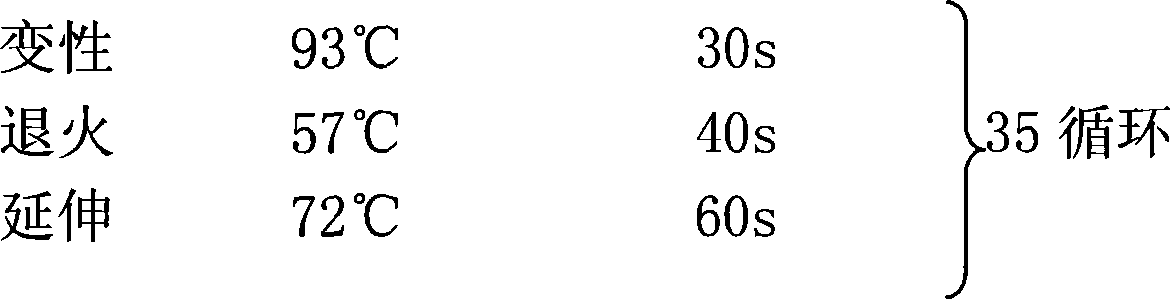Method for processing DNA mould and improving DNA expansion efficiency by using restriction enzymes
A technology of restriction endonuclease and amplification efficiency, applied in the field of using restriction endonuclease to treat DNA template to improve DNA amplification efficiency, to solve the problem of low PCR amplification efficiency, obvious amplification effect, and difficult amplification. increased effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
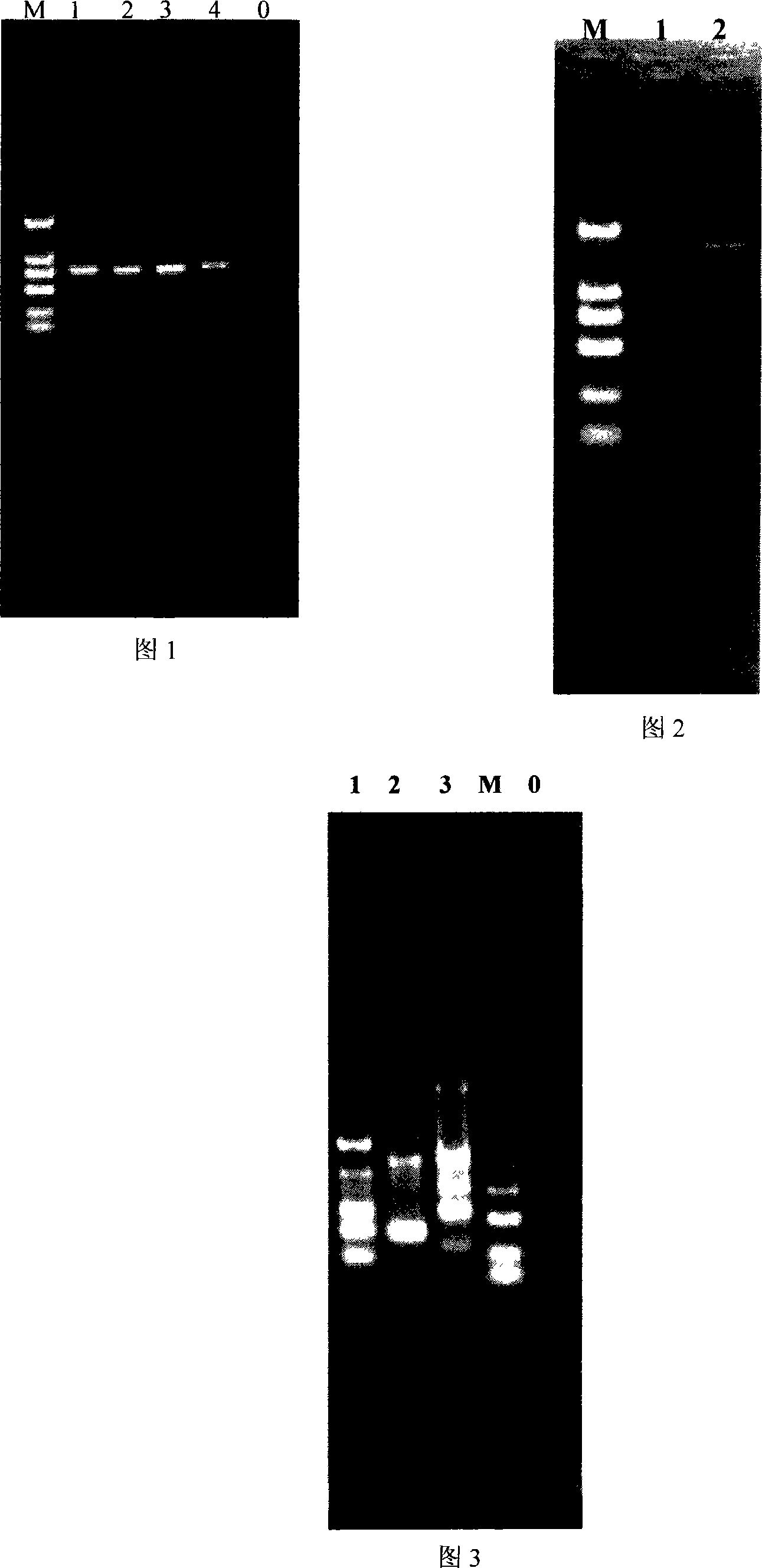
[0052] After digestion with BamHI, BglII, EcoRI, and EcoRV, the amplified fragment is a DNA fragment of 754bp with a GC content of 61%.
[0053] 1. Configuration of PCR reaction system:
[0054] Configure the system in a thin-walled PCR tube in the following order and dosage.
[0055] 10×PCR buffer (with Mg 2+ ) 2.5μL
[0056] dNTP substrate (2.5mM) 2.5μL
[0057]Primer 1 (2.0μM) 2μL
[0058] Primer 2 (2.0μM) 2μL
[0059] Template 2 μL
[0060] Taq enzyme (5U / μL) 0.5μL
[0061] h 2 O up to 25μL
[0062] Among them, the human Genomic DNA template was digested by the above-mentioned various endonucleases, wherein the endonuclease was purchased from Canada BBI Company, and the Taq enzyme was purchased from Beijing Sanbo Yuanzhi Bioengineering Company.
[0063] A total of 5 PCR system tubes were configured, numbered from 0 to 4.
[0064] 2. The templates added sequentially from the 0th to the 4th tubes are undigested and digested with BamHI, BglII, EcoRI, and EcoRV.
[...
Embodiment 2
[0073] The amplified fragments were 790bp DNA fragments with a GC content of 78% after single digestion with BamHI and XhoI respectively.
[0074] 1. Configuration of PCR reaction system:
[0075] Configure the system in a thin-walled PCR tube in the following order and dosage.
[0076] 10×PCR buffer (with Mg 2+ ) 2.5μL
[0077] dNTP substrate (2.5mM) 2.5μL
[0078] Primer 1 (2.0μM) 2μL
[0079] Primer 2 (2.0μM) 2μL
[0080] Template 2 μL
[0081] Taq enzyme (5U / μL) 0.5μL
[0082] h 2 O up to 25μL
[0083] Among them, the human Genomic DNA template was digested by the above-mentioned various endonucleases, wherein the endonuclease was purchased from Canada BBI Company, and the Taq enzyme was purchased from Beijing Sanbo Yuanzhi Bioengineering Company.
[0084] A total of 2 PCR system tubes were configured, and numbered from 1 to 2.
[0085] Note: The template has not been digested, and various measures (optimizing conditions, adding synergists, etc.) still cannot be ...
Embodiment 3
[0095] After the templates were digested with XbaI, EcoRI, and EcoRI+XhoI, the amplified fragments were 1541bp DNA fragments with a GC content of 75%.
[0096] 1. Configuration of PCR reaction system:
[0097] Configure the system in a thin-walled PCR tube in the following order and dosage.
[0098] 10×PCR buffer (with Mg 2+ ) 2.5μL
[0099] dNTP substrate (2.5mM) 3μL
[0100] Primer 1 (2.0μM) 2.5μL
[0101] Primer 2 (2.0μM) 2.5μL
[0102] Template 2 μL
[0103] Taq enzyme (5U / μL) 0.5μL
[0104] Pfu enzyme (2.5U / μL) 0.5μL
[0105] h 2 O up to 25μL
[0106] Among them, the human Genomic DNA template template is digested by the above-mentioned various endonucleases, wherein the endonucleases are purchased from Canada BBI Company, and Taq enzyme and Pfu enzyme are purchased from Beijing Sanbo Yuanzhi Bioengineering Company.
[0107] A total of 4 tubes of the PCR system were configured and numbered from 0 to 3.
[0108] Note: The template has not been digested, and vari...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com