Primer, method and kit for detecting whole exon mutation of SMAD4 gene
An all-exon and exon technology, which is used in biochemical equipment and methods, recombinant DNA technology, and microbial determination/inspection to ensure accuracy and uniqueness, improve risk assessment, and simplify the method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] The primer sequence for detecting SMAD4 gene full-exon mutation includes forward and reverse primers that amplify and cover SMAD4 full-exon mutation site, and its base sequence is:
[0089] SMAD4-1F: CAGGTGTGTCGTAGGATTCG
[0090] SMAD4-1R: GGGAGAAGGTGGCTAGGTTG
[0091] SMAD4-2F: TTTCCTTGCAACGTTAGCTG
[0092] SMAD4-2R: GGAGCACAAATTAAATTACCCTG
[0093] SMAD4-3F: CTGAGTTGGTAGGATTGTGAGG
[0094] SMAD4-3R: TGAAACACTATTGAGATCCTTTTCC
[0095] SMAD4-4F: GCGTTTATGCTACTTCTGAATTG
[0096] SMAD4-4R: TTAATGTTACTGCCTGCCGC
[0097] SMAD4-5F: CCGCTGAATAAATGACTTTTGC
[0098] SMAD4-5R: TTCCAAGTGATTGTGCATACC
[0099] SMAD4-6-7F: CCCATCTTTTATAGTTGTGCATTATC
[0100] SMAD4-6-7R: AAAACAGAAAACAAAGCCCTACC
[0101] SMAD4-8F: TTGGCAGATAGCACTGAAATG
[0102] SMAD4-8R: CCCCATAATTCCATTAAAGCC
[0103] SMAD4-9F: TGTGGAGTGCAAGTGAAAGC
[0104] SMAD4-9R: TGTACATGGGAAAACATAACCTTG
[0105] SMAD4-10F: GAATTCATACTACATGCTCCTGACAC
[0106] SMAD4-10R:TTTTCCATTCCTTCCACCCC
[0107] SMAD4-11F: TCCAAG...
Embodiment 2
[0116] The kit for detecting SMAD4 gene mutations includes: a detection system PCR amplification reaction solution and a sequencing system reaction solution, wherein the detection system PCR amplification reaction solution includes: 2×PCR Buffer; dNTPs (2mM); KOD FX DNA Polymerase (1U / μl); the upper and lower primers SMAD4-1F (10 μM), SMAD4-1R (10 μM), SMAD4-2F (10 μM), SMAD4-2R (10 μM), SMAD4-3F (10 μM), SMAD4-3R(10μM), SMAD4-4F(10μM), SMAD4-4R(10μM), SMAD4-5F(10μM), SMAD4-5R(10μM), SMAD4-6-7F(10μM), SMAD4-6-7R( 10μM), SMAD4-8F(10μM), SMAD4-8R(10μM), SMAD4-9F(10μM), SMAD4-9R(10μM), SMAD4-10F(10μM), SMAD4-10R(10μM), SMAD4-11F(10μM ), SMAD4-11R (10 μM), SMAD4-12F (10 μM), SMAD4-12R (10 μM).
[0117] The sequencing system reaction solution includes: sequencing purification solution (exonuclease I: 0.6U, calf small intestine alkaline phosphatase: 1.2U); EDTA (125mM); absolute ethanol; 75% ethanol; HIDI (highly deionized formamide ); a pair of sequencing primers M13F (3.2 μM), ...
Embodiment 3
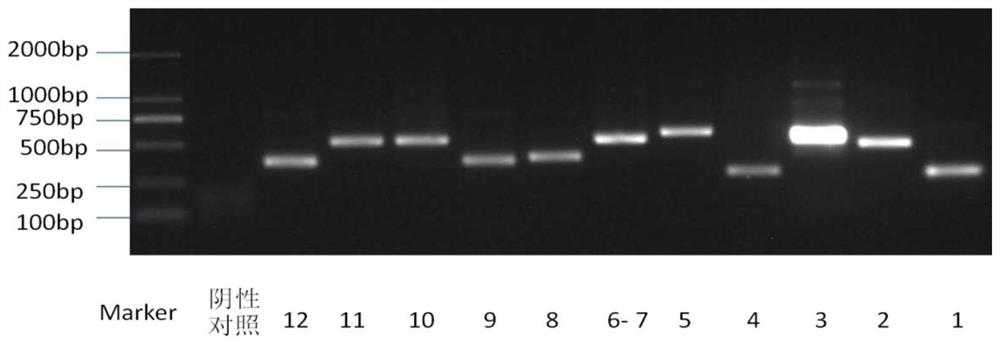
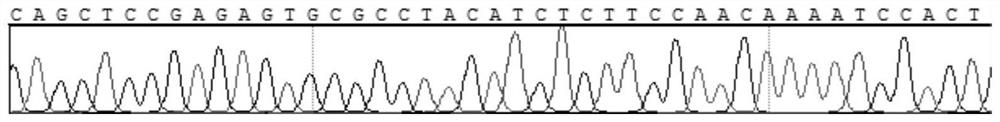
[0121] Method for detecting SMAD4 gene mutation site
[0122] (1) DNA extraction from tissue samples
[0123] Extract DNA from human tissue samples (using QIAamp DNA FFPE TissueKit, a DNA extraction kit from Qiagen), the specific extraction method is as follows:
[0124] 1) Remove the non-cancerous tissue with a razor blade, collect the paraffin-embedded tissue into a 1.5mL centrifuge tube, and centrifuge briefly.
[0125] 2) Add 1mL tissue clear solution, shake fully, centrifuge at 13200rpm for 1min, and discard the supernatant.
[0126] 3) Add 0.5mL tissue clear solution, shake fully, centrifuge at 13200rpm for 1min, and discard the supernatant.
[0127] 4) Add 1 mL of absolute ethanol, shake vigorously, centrifuge at 13200 rpm for 2 min, and discard the supernatant.
[0128] 5) Open the lid and incubate at room temperature or in a metal bath at 37°C until the ethanol evaporates completely.
[0129] 6) Take 180 μL Buffer ATL to resuspend the tissue in the tube, add 20 μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com