Protein folding optimizing method based on ant group algorithm
A technology of protein folding and optimization method, applied in the field of protein folding optimization using ant colony algorithm, to achieve the effect of fast and effective solution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
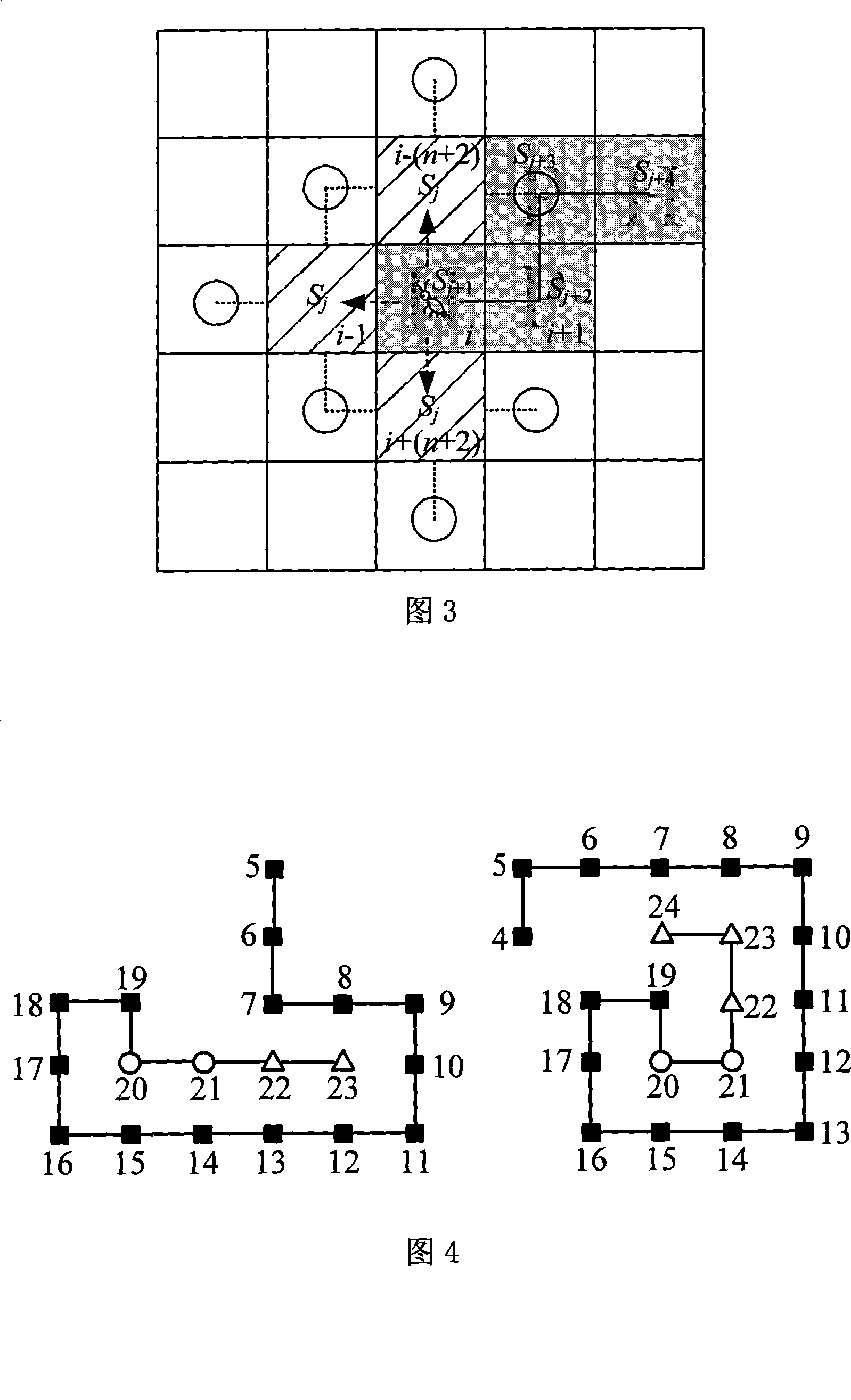
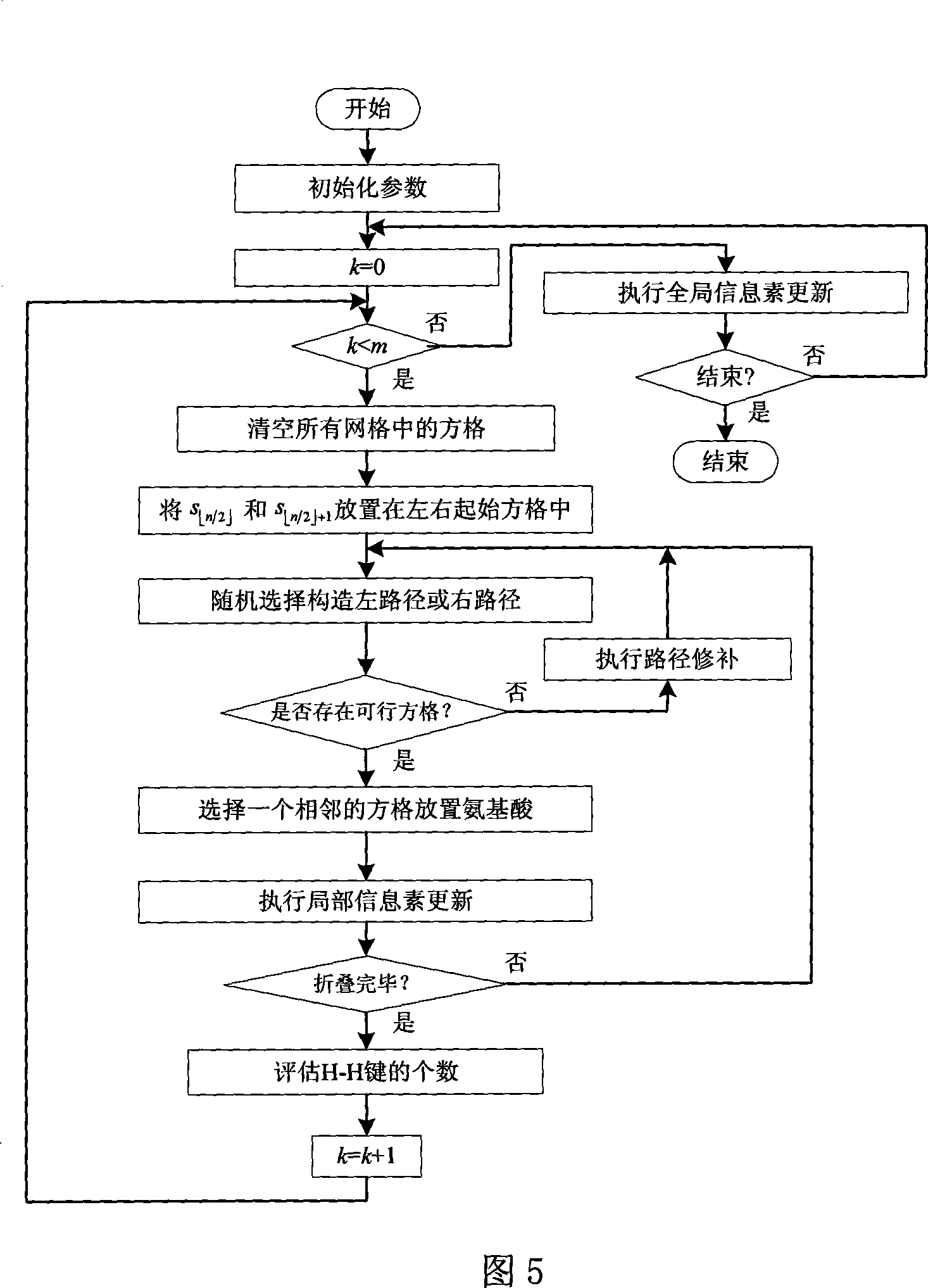
[0034] The method of the invention will be further described below in conjunction with the accompanying drawings.
[0035] There are a total of twenty amino acids in nature. Using different classification methods, they can be classified as acidic, basic or neutral; positively charged, negatively charged or uncharged; hydrophobic or hydrophilic and so on. For globular proteins in aqueous solution, its hydrophilic amino acids tend to distribute on the surface of the sphere due to the attraction of water molecules. Since hydrophobic amino acids are repelled by water molecules, except for some special hydrophobic amino acid regions distributed on the surface, most of the remaining hydrophobic amino acids will gather in the interior of the globular protein to form a core.
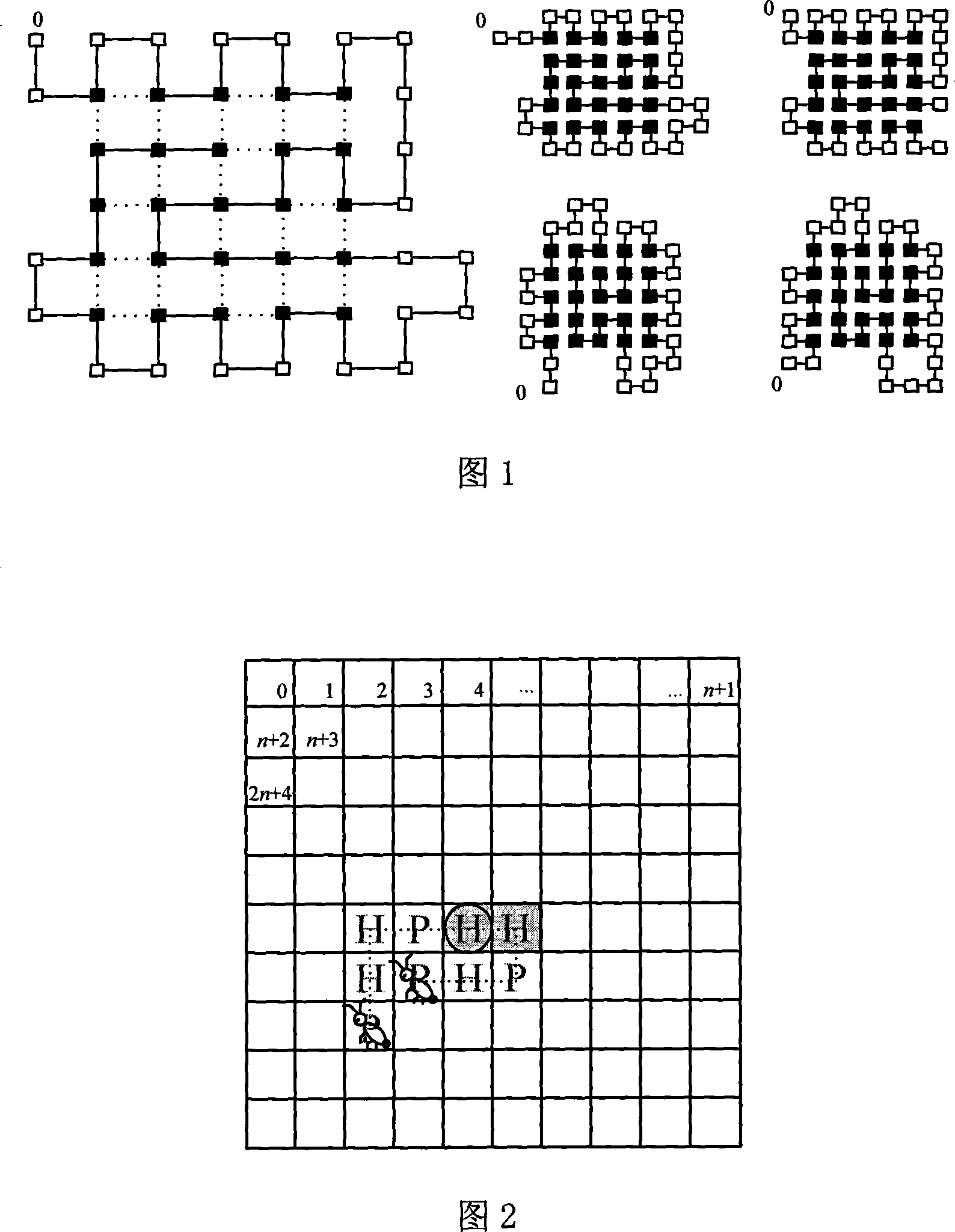
[0036] Some two-dimensional HP fold structures of protein sequences containing 48 amino acids are shown in Fig. 1, where solid squares represent hydrophobic amino acids and open squares represent hydrophilic ami...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com