Cucumber powdery mildew resistance main effect QTL compact linkage molecule labeling method and applying method
A technology of cucumber powdery mildew and molecular markers, which is applied in the directions of biochemical equipment and methods, and microbial determination/inspection, can solve the problem of low disease-resistance coincidence rate in the field, improve breeding and selection efficiency, improve breeding efficiency, and speed up Select the effect of progress
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
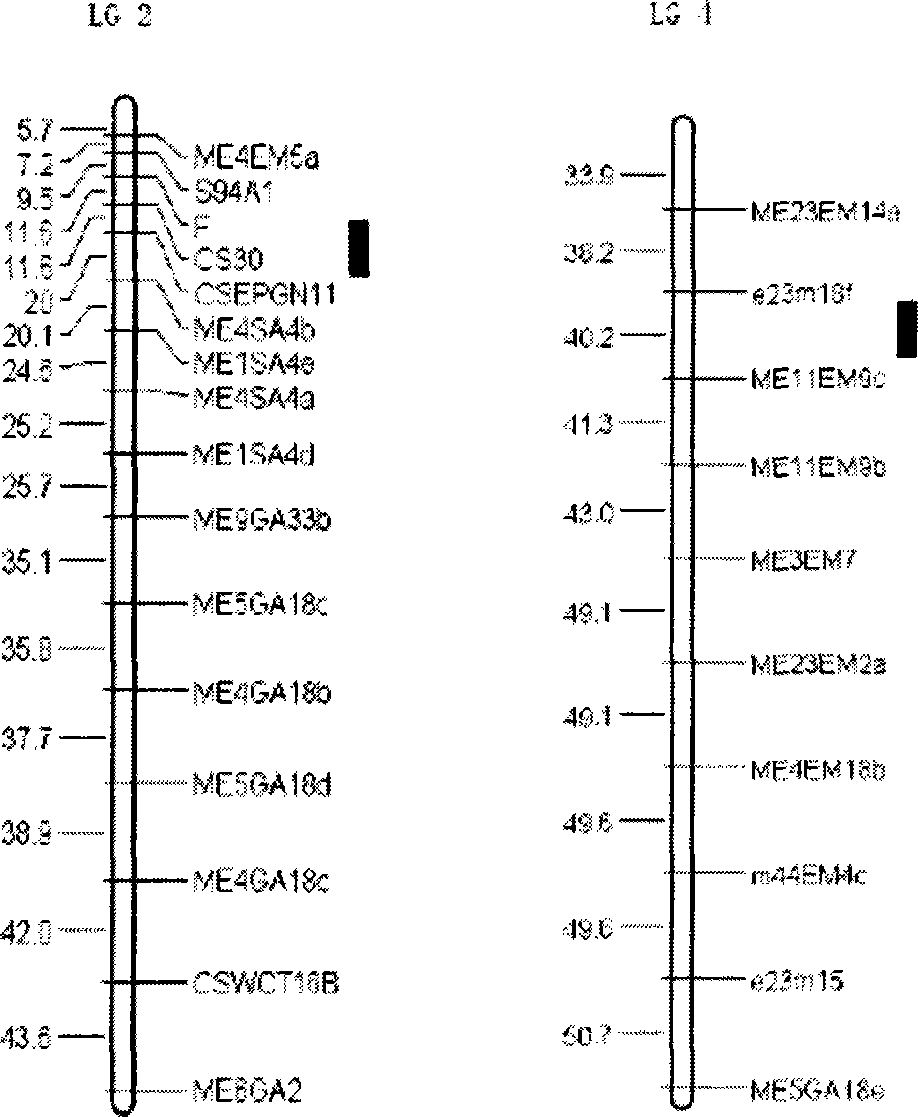
[0026] Molecular marker method of closely linked major QTL for powdery mildew resistance in cucumber
[0027] (1) Using S06 to cross with cucumber powdery mildew susceptible variety S94 to produce F 1 mongrel.
[0028] (2)F 1 F 2 , F 2 The 252 single plants of the 252 plants were randomly selected for planting a seed, and propagated to F 7 generation, resulting in 224 families.
[0029](3) Select SRAP, SSR, SCAR and STS primer combinations with amplified polymorphisms between the two parents, and use existing methods for 224 families (Wang Gang, Chinese Science C Life Science 2004, 34 (6): 510 ~516; Sakata Y, Theor Appl Genet (Applied Genetic Theory), 2006, 112: 243-250) for analysis, isolate the DNA of cucumber leaves of each recombinant inbred line, and use related sequence amplified polymorphism (SRAP), Microsatellite (SSR), sequence-specific amplification region (SCAR) and sequence tag site (STS) molecular marker primers carry out PCR amplification, and the amplified...
Embodiment 2
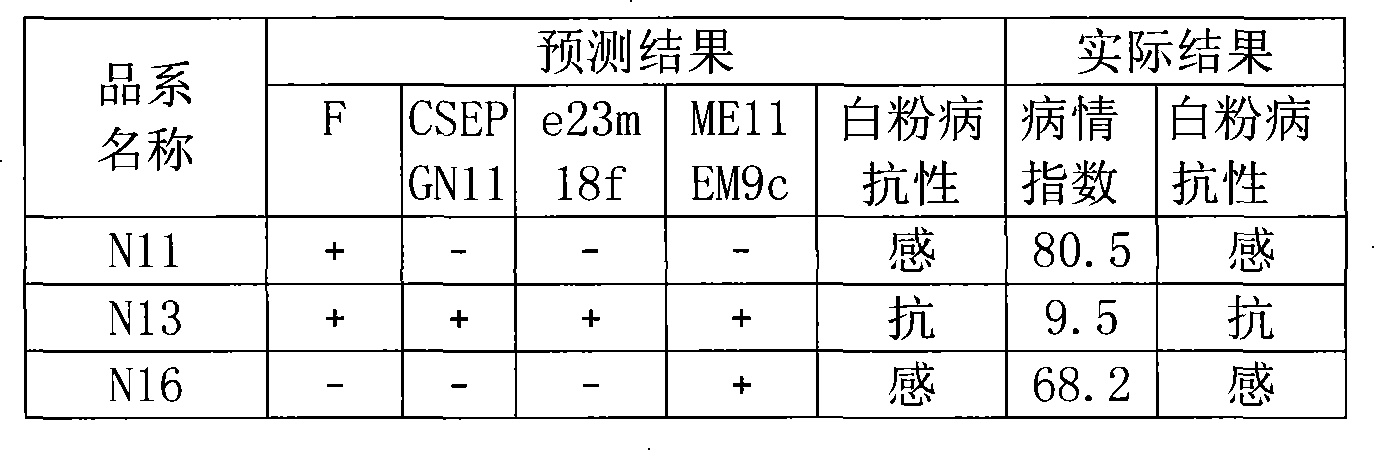
[0041] Validation of molecular markers linked to the main QTL for powdery mildew resistance of S06 in a high-generation genetic population:
[0042] Molecular markers closely linked to the major QTL for powdery mildew resistance in cucumber were obtained. The powdery mildew resistance was predicted for some plants of the F8 offspring crossed between S06 and S94. DNA was isolated from their leaves first, and then the marker F , CSEPGN11, e23m18f, and ME11EM9c primers were used to perform polymerase chain reaction (PCR) on these DNAs, and determine whether there were corresponding markers by judging the maps. If there were more than 3 molecular markers, it indicated that the variety or strain belonged to powdery mildew resistant Sexual material, its absence is susceptible material. The powdery mildew resistance of each cucumber plant was then predicted based on these criteria. Then, the actual powdery mildew resistance of the tested samples was measured by inoculating the powde...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com