Method for detecting A1762T-G11764A double-mutation of hepatitis B virus DNA and kit
A technology of DNA A1762T-G1764A and A1762T-G1764A, which is applied in the field of detection of hepatitis B virus DNA A1762T-G1764A double mutation, can solve the problems of linear probe specificity not as good as molecular beacon and troublesome sequencing method, and achieve beautiful detection results And stable, reliable test results, conducive to the effect of popularization and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Embodiment 1: Extraction of hepatitis B virus DNA in clinical blood samples
[0078] Take 50 μl of serum from clinical blood samples into a high-pressure 1.5ml centrifuge tube, add 25 μl of extraction solution A (40% PEG-8000), shake and mix for 5 seconds, centrifuge at 12,500 g for 5 minutes; discard the supernatant, add 25 μl of extraction Solution B (0.25M NaOH), shake for 15s to dissolve the precipitate as much as possible, lyse at 96°C for 10min; add 25μl of extract solution C (0.4M Tris-HCl buffer, pH6.4), mix well, 96°C for 10min; centrifuge at 12,500g After 15 minutes, 5 μl of the supernatant was taken for PCR amplification.
Embodiment 2
[0079] Embodiment 2: PCR amplification of target nucleic acid
[0080] Add a single serving of PCR reaction mixture into a PCR reaction tube, then add 0.2 μl of enzyme mixture, and finally add 5 μl of template (template to be tested or negative and positive control), mix thoroughly and centrifuge slightly (about 5 Second). Then put the reaction tube into a PCR machine (a PCR machine that can detect FAM), and amplify according to the following conditions: 50°C for 120 seconds (1 cycle) → 95°C for 180 seconds (1 cycle) → 95°C for 10 seconds, 58°C °C for 30 seconds, 35 °C for 30 seconds (40 cycles). Among them, 58°C is the annealing temperature, and 35°C is the temperature for detecting fluorescence.
[0081] The PCR amplification system used included: 2.5 μl 10×PCR buffer (850 mM KCl, 400 mM Tris-Cl, pH8.9, 50% v / v glycerol), 0.2 μl enzyme mixture, 0.2 μl dUTPs (containing dATP, dUTP , dGTP, dCTP each 25mM), two forward and reverse primers (SEQ ID NO: 1 and SEQ ID NO: 2, 100 ...
Embodiment 3
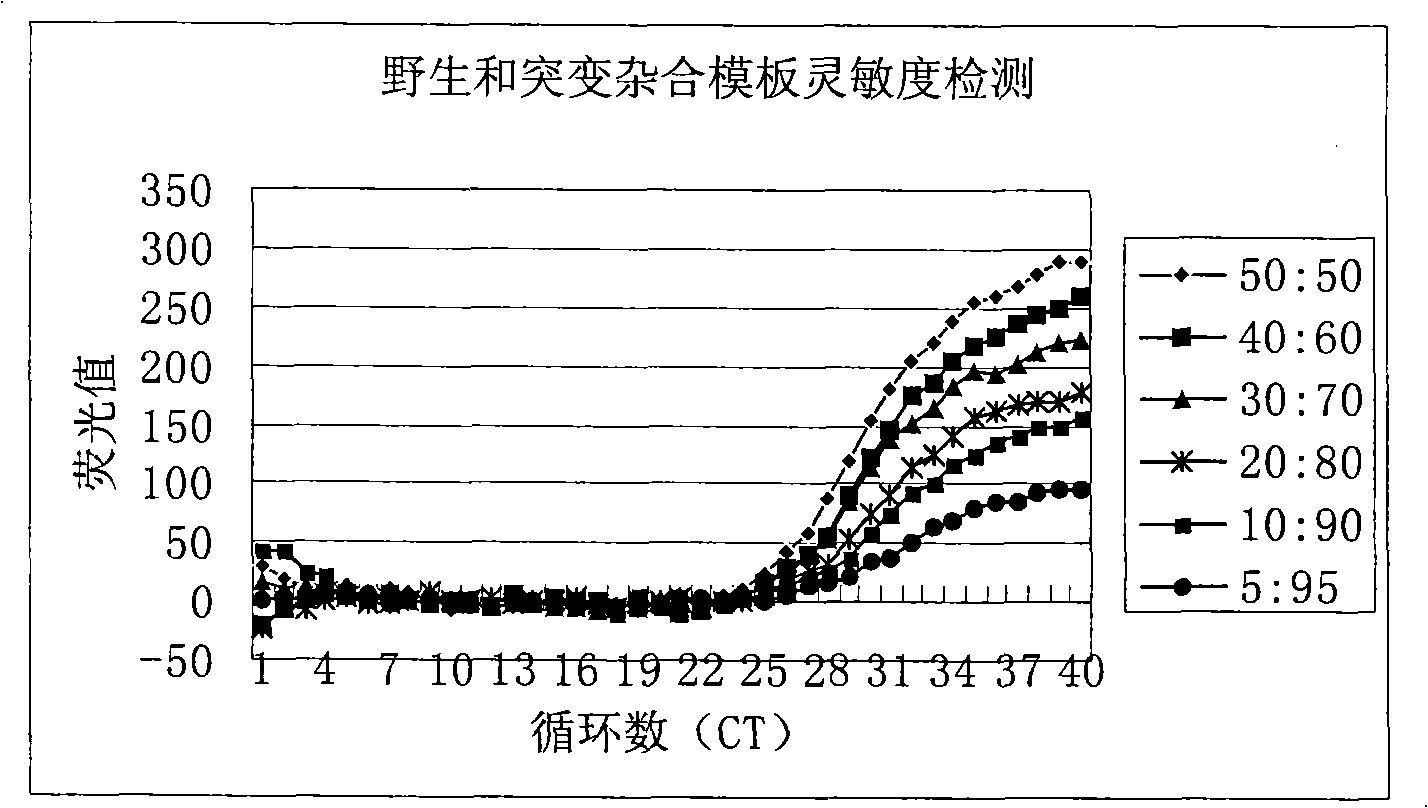
[0082] Example 3: Sensitivity and specificity detection of hepatitis B virus DNA A1762T-G1764A double mutation
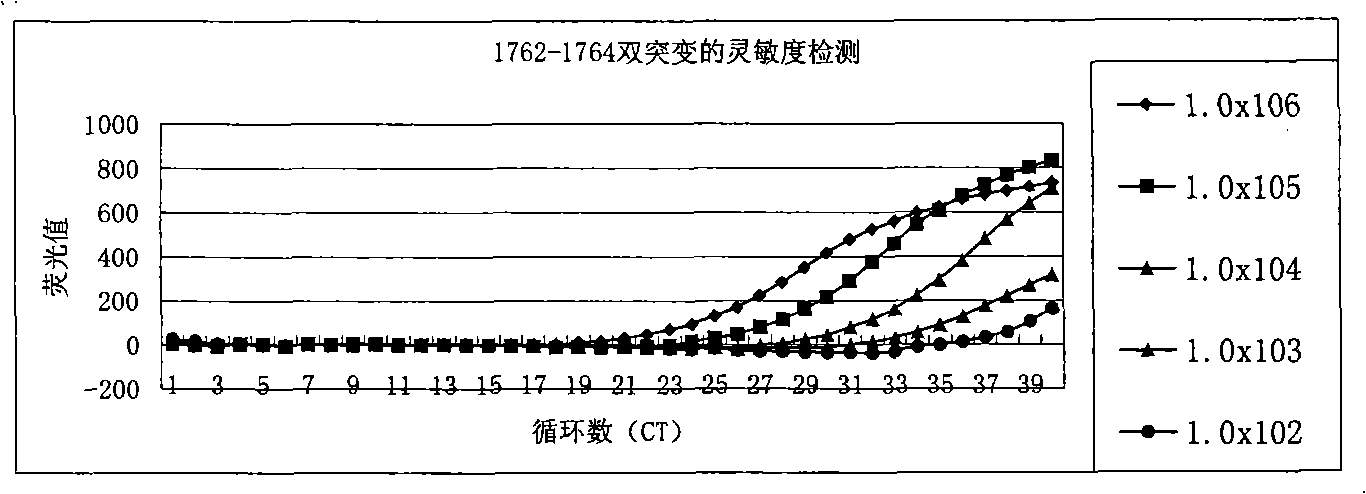
[0083] The plasmid (1.0×10 6 copies / ml-1.0×10 2 copy / ml) is template, use reagent of the present invention to hepatitis B virus DNA A 1762 T-G 1764 The sensitivity of the A double mutation was detected, and the detection results can be found in the appendix figure 1 . From figure 1 As can be seen from the results, the method and kit of the present invention can at least detect that the template concentration is only 1.0×10 2 copy / ml of A 1762 T-G 1764 A double mutant hepatitis B virus DNA. In other words, the sensitivity of the present invention can fully meet the clinical needs.
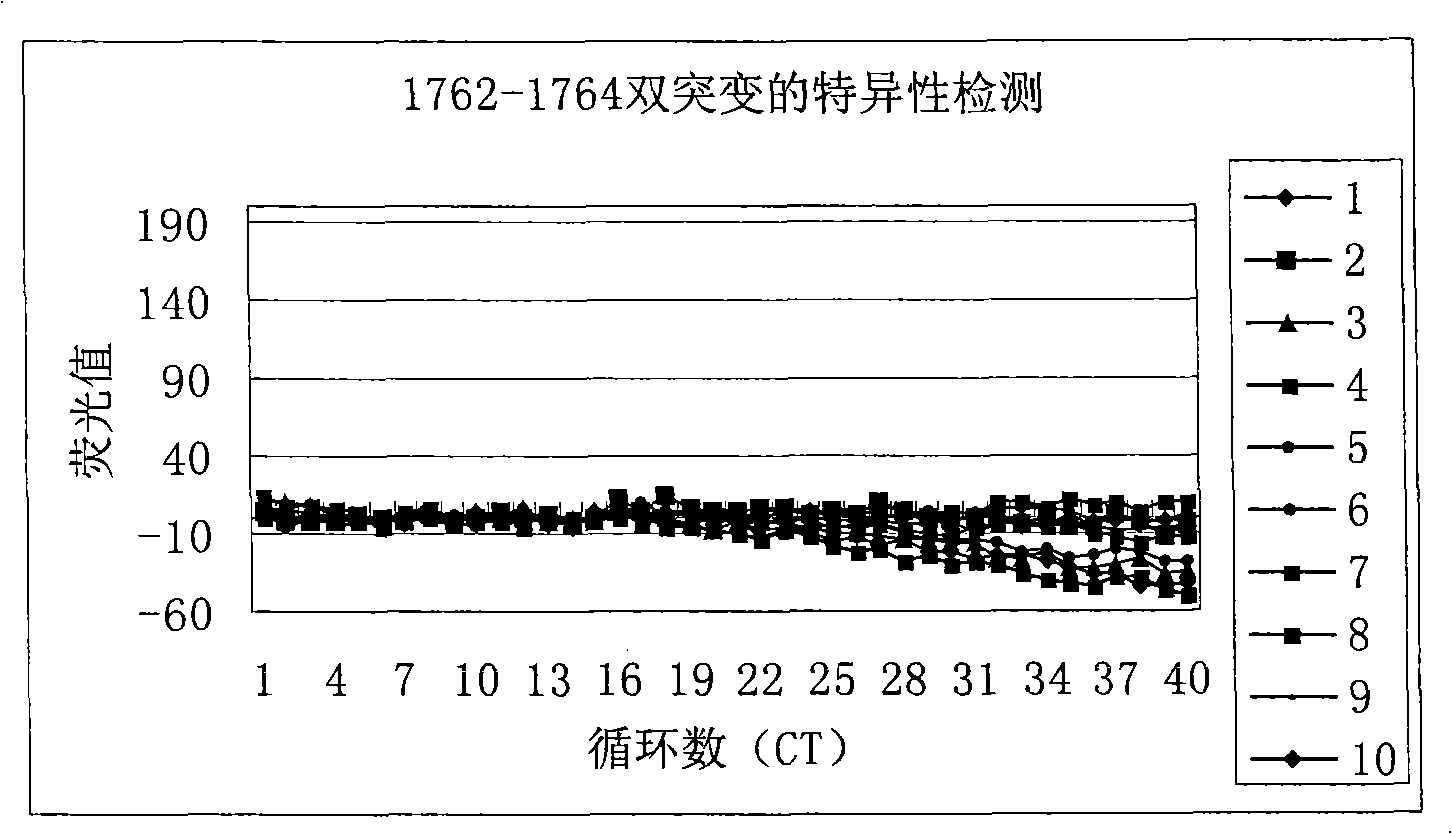
[0084] With 10 plasmids (approximately 1.0 × 10 6 copies / ml) is template, use reagent of the present invention to hepatitis B virus DNA A 1762 T-G 1764 The specificity of the A double mutation was tested, and the test results can be found in the appendix figure 2 . From f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com