Signal combination coding-based DNA ligation sequencing method
A DNA sequencing and signal combination technology, applied in the biological field, can solve the problems of single signal labeling method, low efficiency, sequencing cost, throughput and speed that cannot meet the needs of life sciences, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
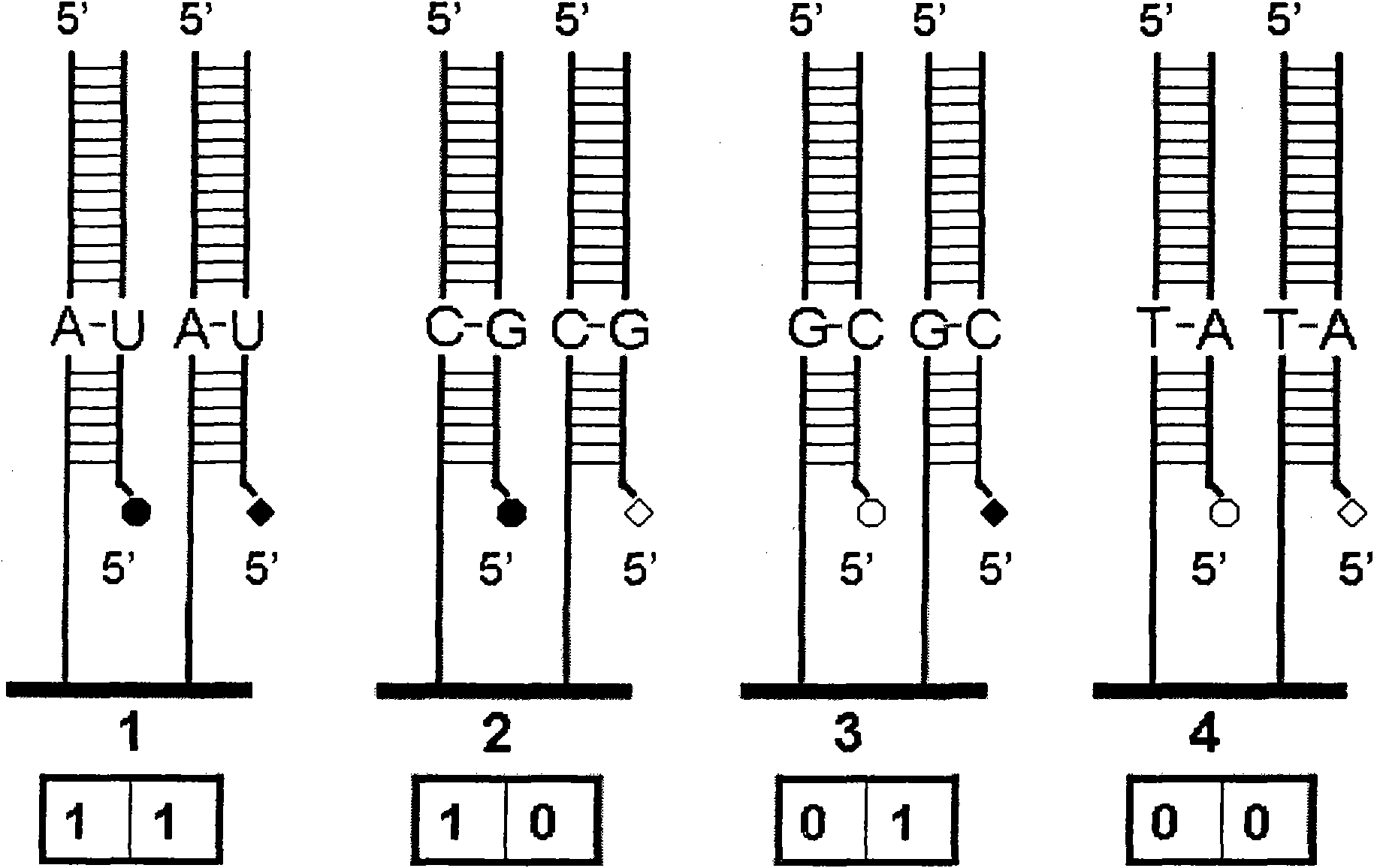
[0047] Example 1: The present invention is a DNA ligation sequencing method based on signal combination coding, which is characterized in that a specific DNA sequencing probe is labeled with a combination of multiple marker states, and a batch of DNA sequencing probes are used for labeling. The needle is labeled with different combinations of multiple marker states to prepare a set of DNA sequencing probes encoded by the signal combination, so as to distinguish and identify different DNA sequencing probes during detection, so as to achieve the goal of sequencing the template to be tested. purpose.
[0048]A. Preparation of DNA sequencing probes encoded by a set of signal combinations: first prepare unlabeled DNA sequencing probes. Each unlabeled DNA sequencing probe consists of one or more sequencing bases and one or more degenerate bases. Base N or non-strictly paired base composition. Sequencing bases are used to determine the base information of the corresponding position in th...
Embodiment 2
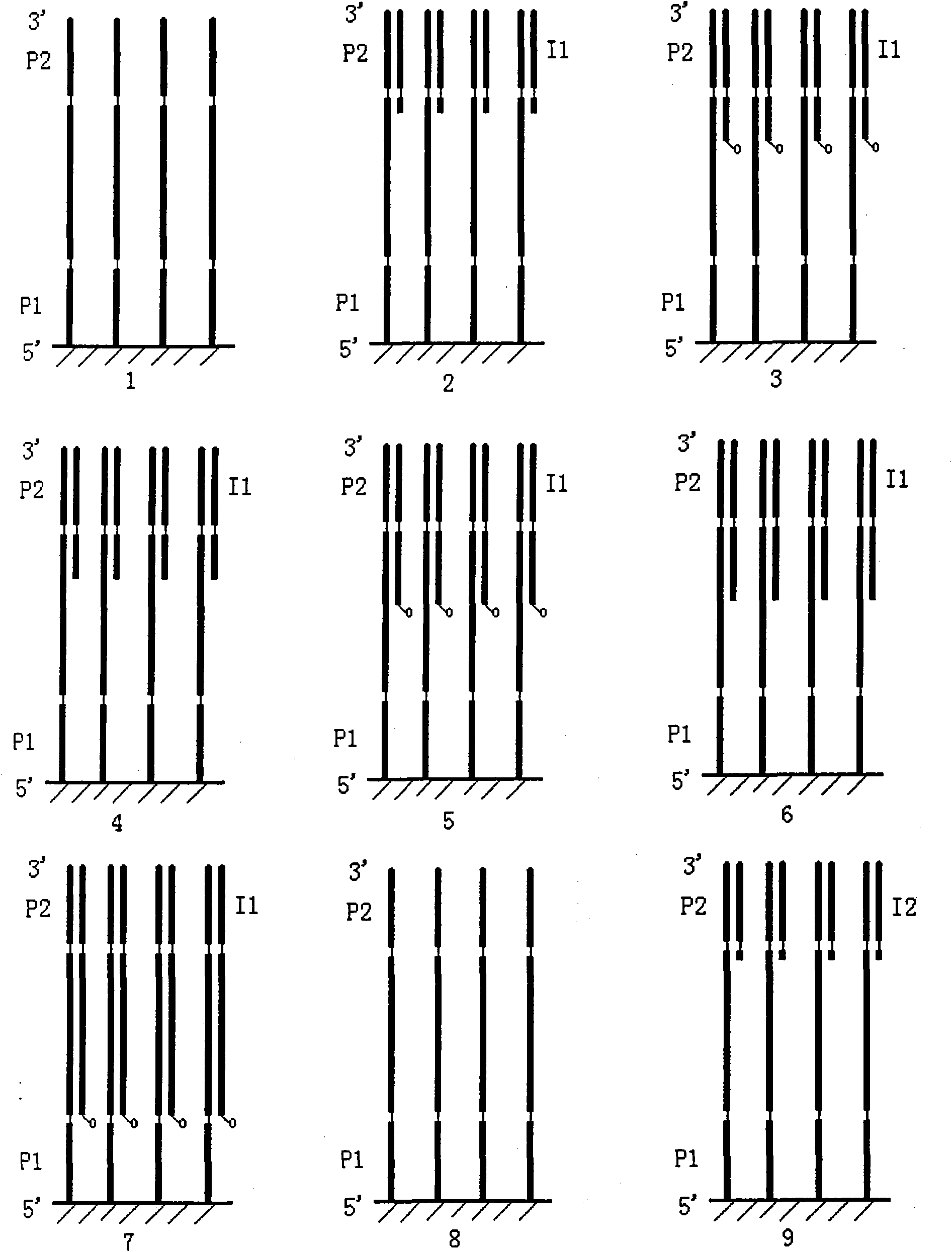
[0065] Example 2: Four-color fluorescence combination coding method (respective labeling scheme) detects a total of 32 base pair sequences of 45332-45363 on human chromosome 17 on the RP11-354P11 clone;
[0066] Extract human whole genome DNA, use PCR method to amplify the target fragment, the primers are P1, P2 (see Table 3 for the sequence), and the 5'end of primer P1 is modified with hydroxyl group. After the amplified PCR product is fixed on the surface of the aldehyde-modified glass slide, the glass slide is heated to 95°C to separate one strand of the double-stranded PCR product from the fixed strand, and then the sequencing reaction is started.
[0067] Table 3 Related oligonucleotide sequence information in Example 2
[0068] Sequence name
Sequence information
Sequence to be tested
5’-CACGGACCAGCTGCCCTGGACCAGCTGCAAGA-3’
P1
OH-5’-CGCTATACTACCTCATCTCCTCCTTCACG-3’
P2
5’-GCAGTTGCCAGTGTTCCAGGAGT-3’
I1
5’-GTTCCAGGAGT...
Embodiment 3
[0077] Example 3: Four-color fluorescence combination coding method (common labeling scheme) detects a total of 32 base pair sequences of 45332-45363 on human chromosome 17 on the RP11-354P11 clone;
[0078] Extract human whole genome DNA, use PCR method to amplify the target fragment, the primers are P1, P2 (see Table 5 for the sequence), and the 5'end of primer P1 is modified with hydroxyl group. After the amplified PCR product is fixed on the surface of the aldehyde-modified glass slide, the glass slide is heated to 95°C to separate one strand of the double-stranded PCR product from the fixed strand, and then the sequencing reaction is started.
[0079] Table 5 Related oligonucleotide sequence information in Example 3
[0080] Sequence name
Sequence information
Sequence to be tested
5’-CACGGACCAGCTGCCCTGGACCAGCTGCAAGA-3’
P1
OH-5’-CGCTATACTACCTCATCTCCTCCTTCACG-3’
P2
5’-GCAGTTGCCAGTGTTCCAGGAGT-3’
I1
5’-GTTCCAGGAGTNNNN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com