Method for detecting cattle Six6 gene single nucleotide polymorphism
A single nucleotide polymorphism and gene technology, applied in the field of molecular genetics, can solve problems such as blanks and lack of research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
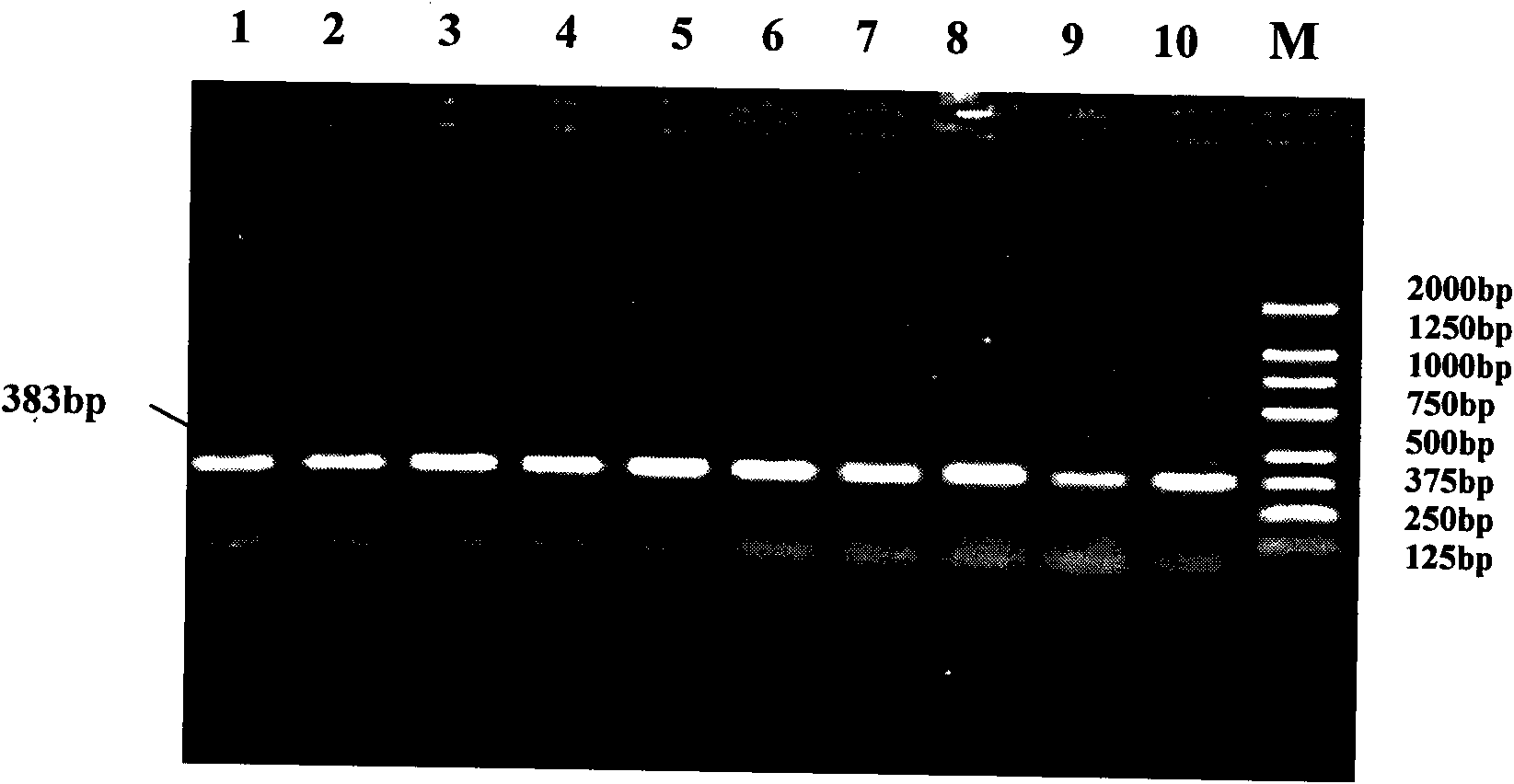
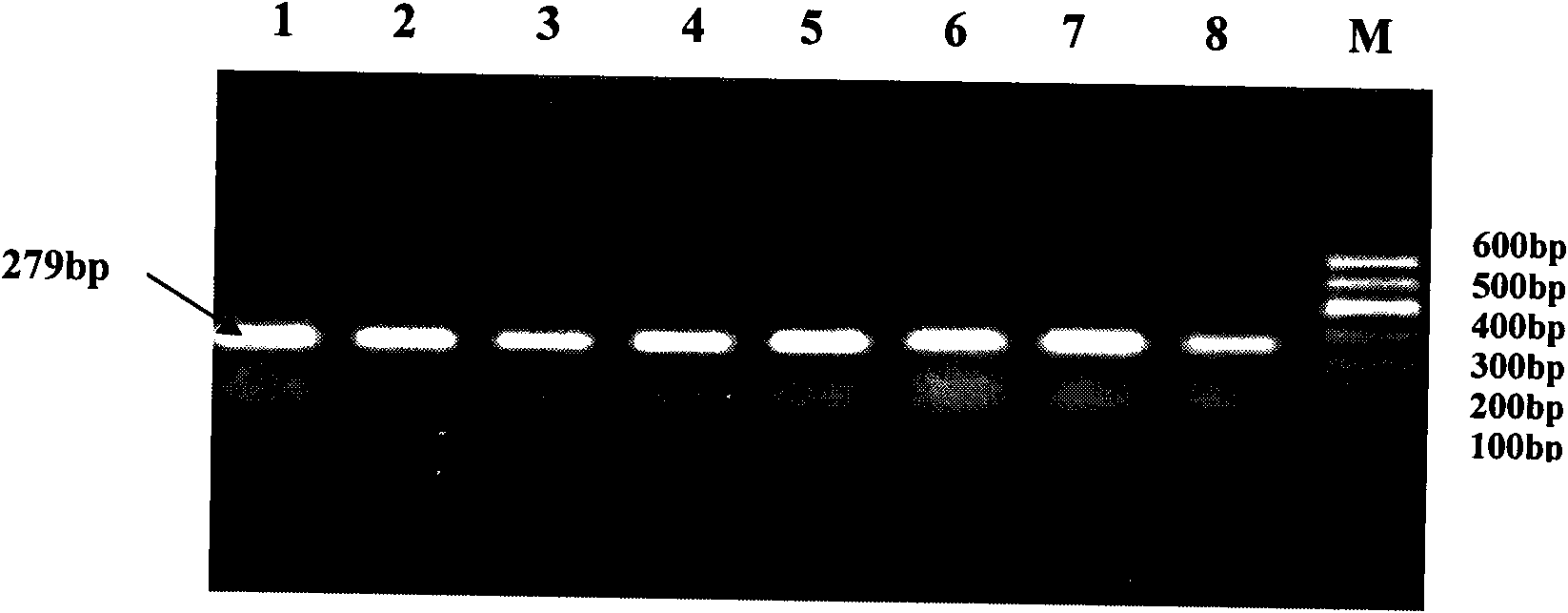
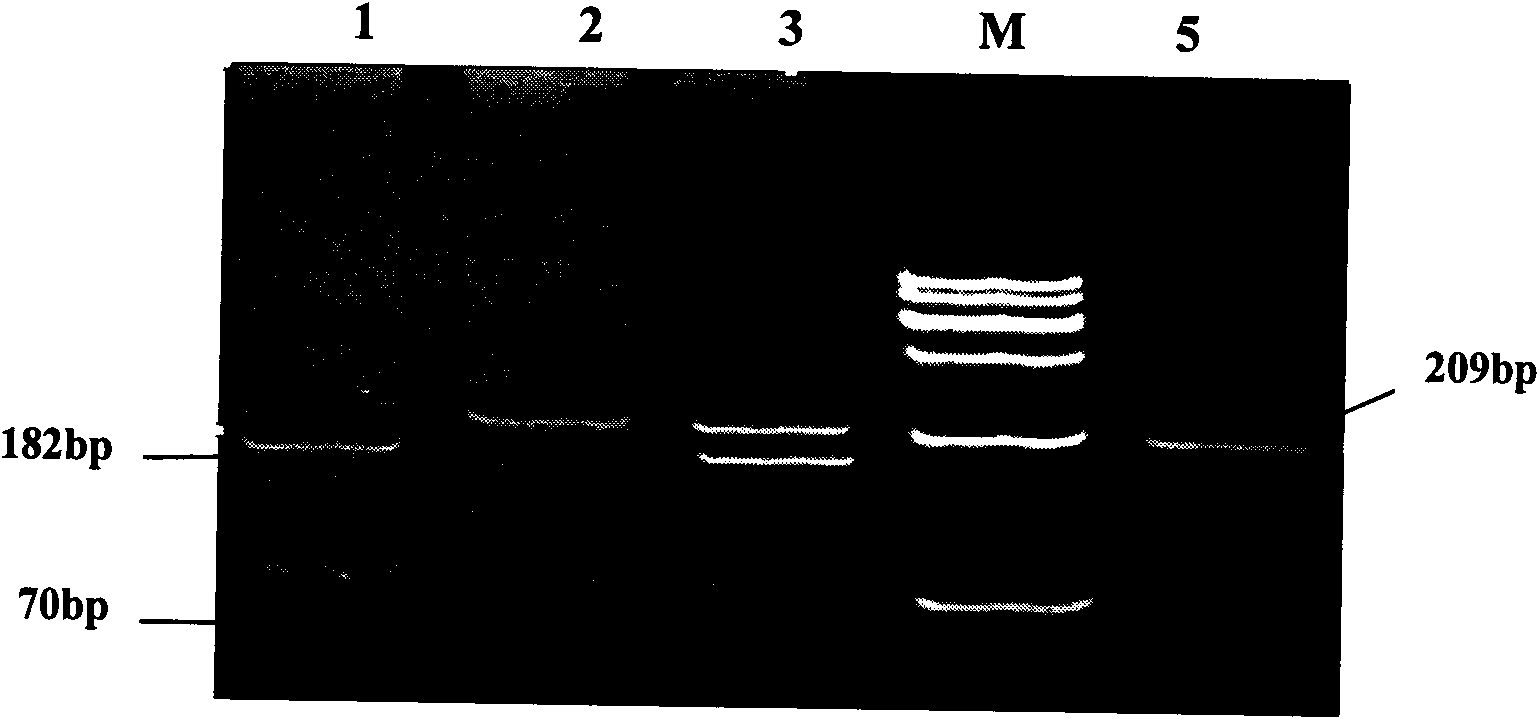
[0026] The present invention uses the RFLP-PCR method to detect the single nucleotide polymorphism that may result in a change in the conformation of the encoded protein due to the mutation of the nonsense codon at position 2015 of the cattle Six6 gene. The present invention will be further described in detail below. They are presented by way of explanation, not limitation, of the invention.
[0027] a. Design of PCR primers for the region containing the stop codon of cattle Six6 gene
[0028] Using the Hereford Cattle (NC_007308) sequence published by NCBI as a reference, Primer 5.0 was used to design PCR primers capable of amplifying the region containing the stop codon of the cattle Six6 gene. The primer sequences are as follows:
[0029]Upstream primer: gggctgactg ctgggctta 19;
[0030] Downstream primer: agaccaagca acccagcg 18;
[0031] Using the above primers to amplify the cattle genome, it is possible to amplify a 383bp gene fragment comprising the stop codon region ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com