Molecule detection method of spongioblast endosymbiotic actinomycetes
A technology for molecular detection and actinomycetes, which is applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., can solve the problem that actinomycetes are not paid enough attention, rarely obtain information on the diversity of spongy actinomycetes and Distribution characteristics, lack of actinomycete species-specific primers, etc., to achieve accurate diversity information and improve sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0016] The technical solutions of the present invention will be further described below in conjunction with the accompanying drawings and embodiments. The following examples are not intended to limit the present invention.
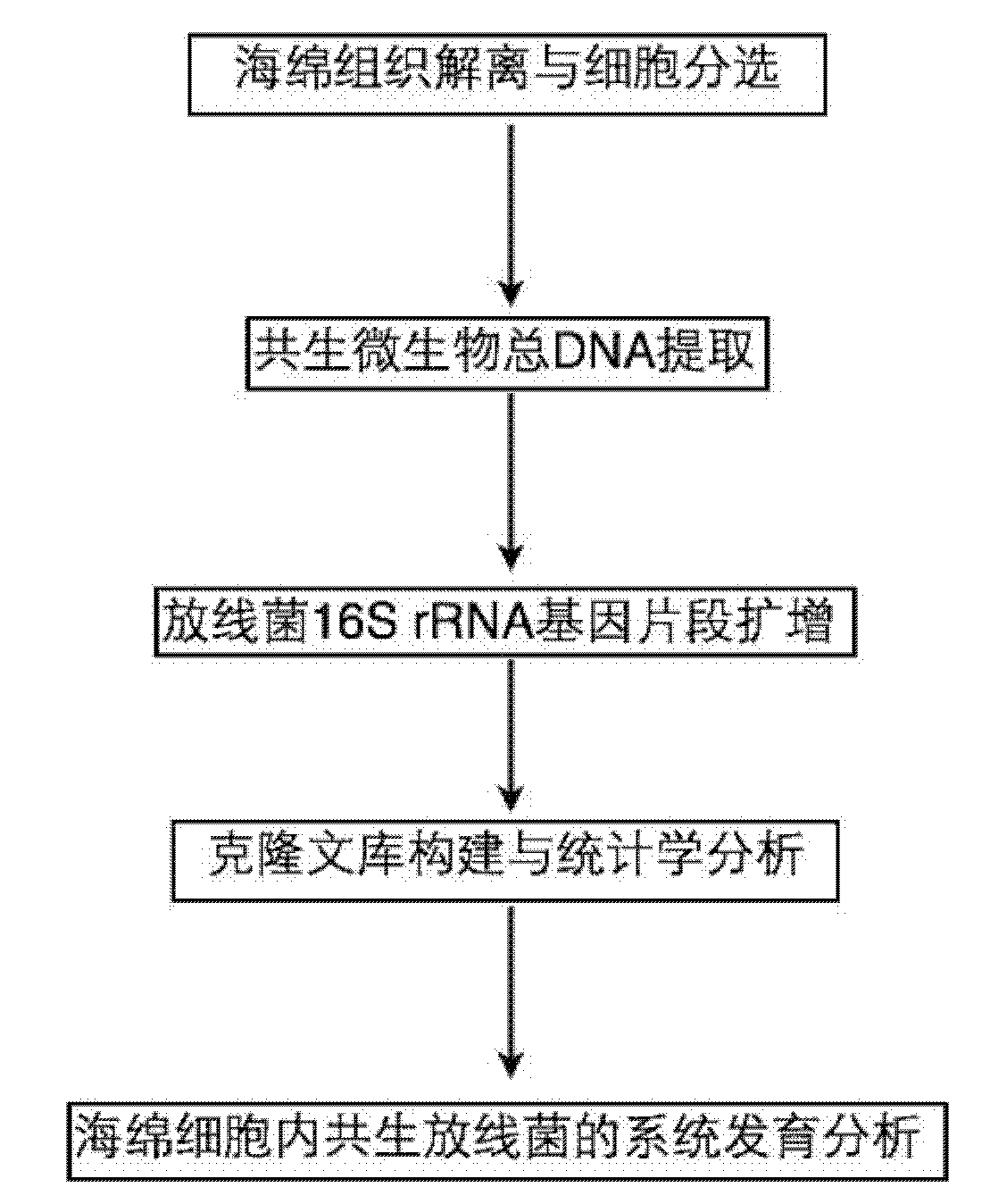
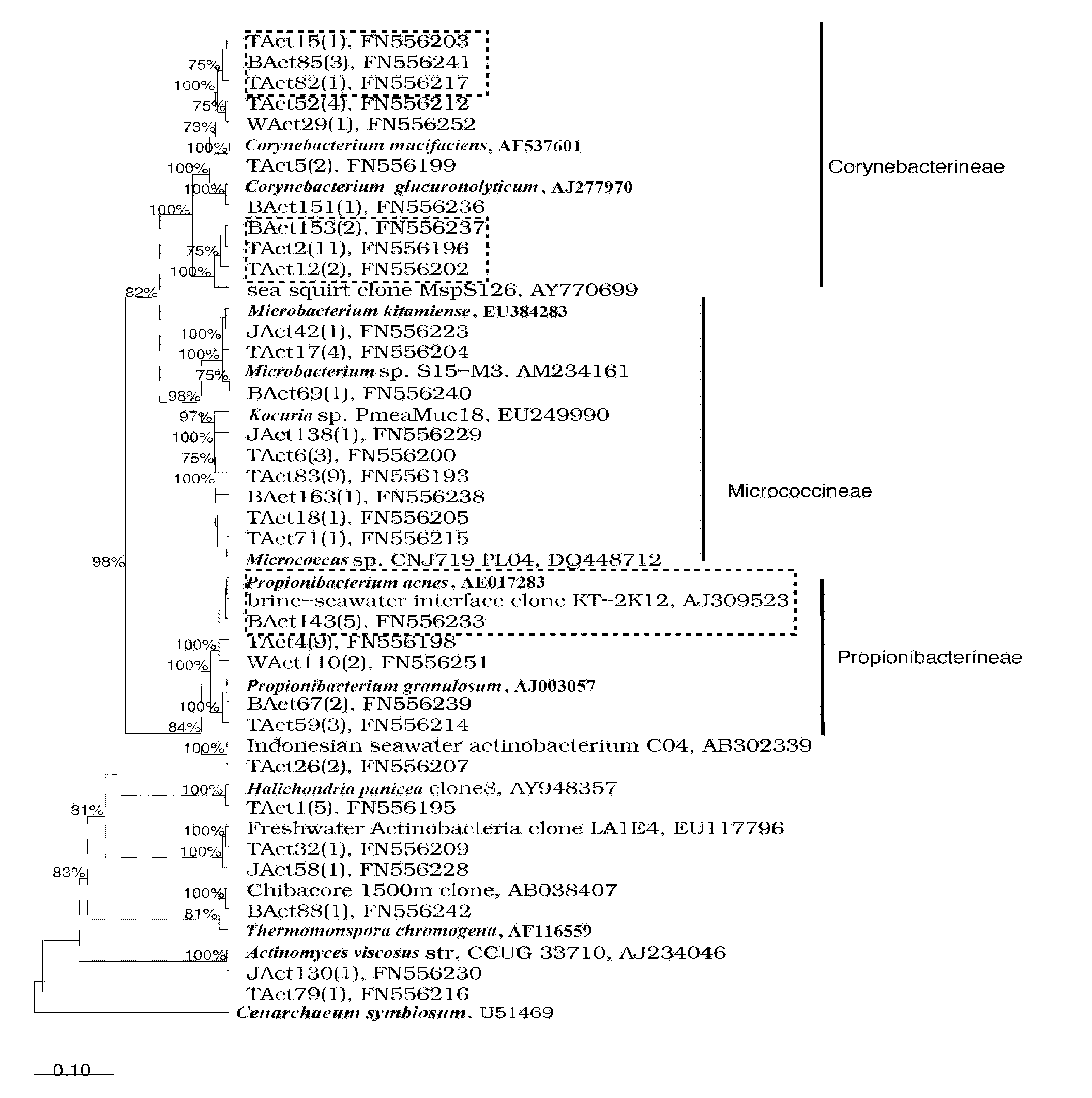
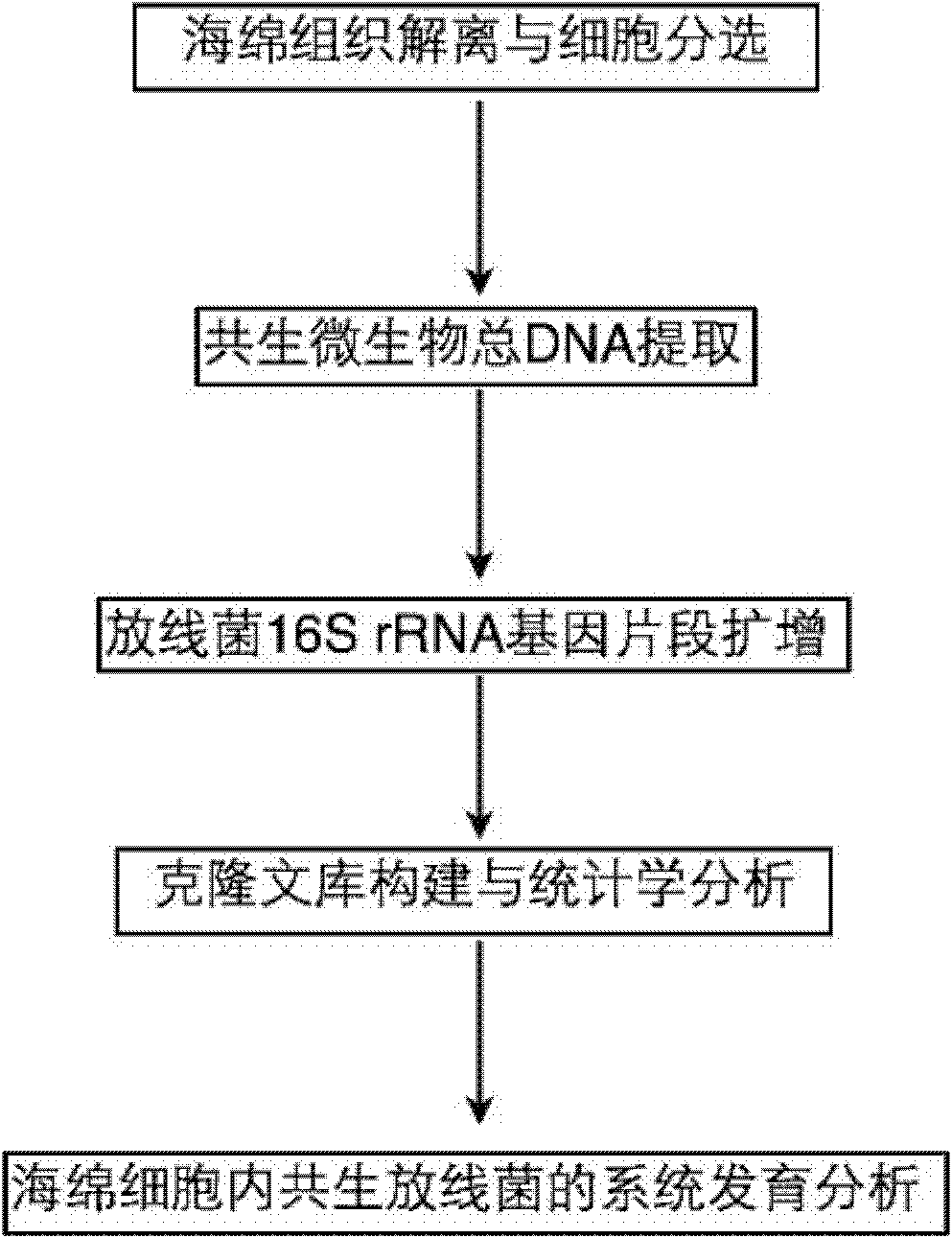
[0017] The flow process of the inventive method is as figure 1 As shown, first dissociate the sponge tissue and sort the cells to obtain the sponge cells, extract the total DNA of the endosymbiotic microorganisms in the cells, amplify the 16S rRNA gene fragments of actinomycetes by polymerase chain reaction, and combine them by constructing a cloning library. Statistical and phylogenetic analysis revealed the composition of symbiotic actinomycetes in sponge cells, and found specific actinomycetes in sponge cells.
[0018] The specific implementation steps are as follows:
[0019] 1. Spongy tissue dissociation and cell sorting:
[0020] (1) The following operations are performed under sterile conditions. Spongy tissue (1-2g) was thawed at 4°C and cut in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com