Method for identifying pure Hareford bulls and hybrid Hareford bulls
A Hereford, purebred technology, applied in the field of animal husbandry, can solve the problem of not identifying the purebred cows of Hereford, and achieve the effects of good social benefits, considerable economic benefits, and strong practicability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
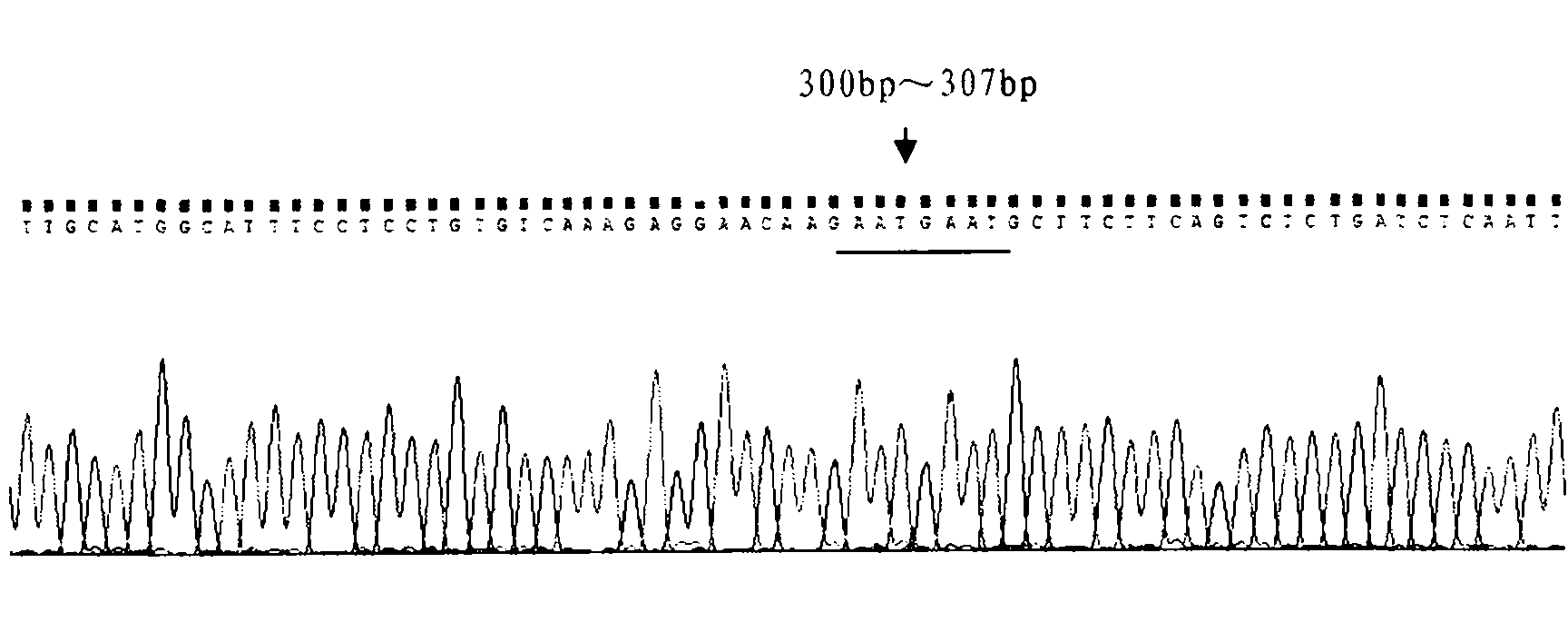
[0021] Genomic DNA was extracted from the first target bovine blood by phenol-chloroform extraction, and specific primers were designed to amplify the TLR2 gene promoter region fragment. The primer sequences used were:
[0022] The upstream primer sequence is: SEQ ID NO.4:
[0023] TLR2-promoter-F: 5'-ggattctccaggcaagaaca-3';
[0024] The downstream primer sequence is: SEQ ID NO.5:
[0025] TLR2-promoter-R: 5'-agtcctgtggtggtcccttt-3'.
[0026] The PCR reaction system used was as follows: the total system was 20 microliters (μl), including 10-100 ng of genomic DNA, 1.0 μl of upstream primers (10 pmol / μl), 1.0 μl of downstream primers (10 pmol / μl), 2.0 μl of 10 ×PCR Buffer (containing magnesium ions Mg 2+ ), 2 μl of dNTPs (0.25 mM), 2.0 U of Taq DNA polymerase and sterilized water.
[0027] The PCR reaction program used was as follows: pre-denaturation at 94°C for 4 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s; e...
Embodiment 2
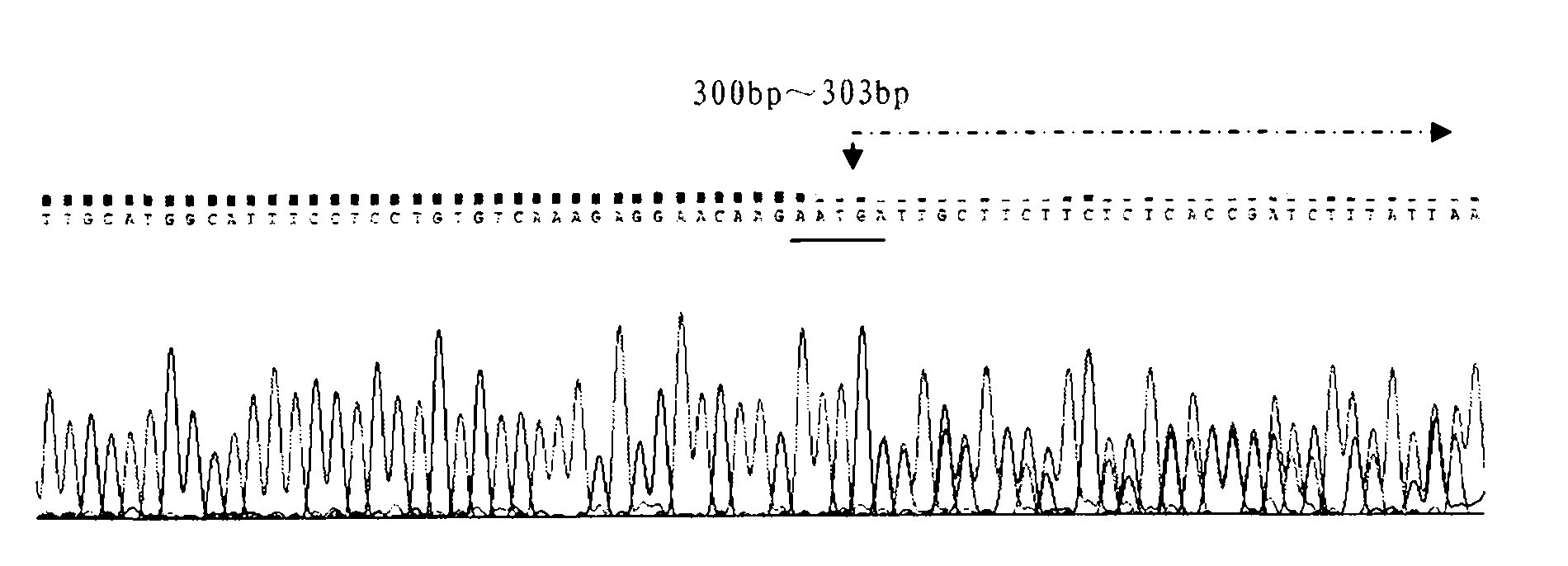
[0030] Genomic DNA was extracted from the second target bovine blood by phenol-chloroform extraction, and specific primers were designed to amplify the TLR2 gene promoter region fragment. The primer sequences used were:
[0031] The upstream primer sequence is: SEQ ID NO.4:
[0032] TLR2-promoter-F: 5'-ggattctccaggcaagaaca-3';
[0033] The downstream primer sequence is: SEQ ID NO.5:
[0034] TLR2-promoter-R: 5'-agtcctgtggtggtcccttt-3'.
[0035] The PCR reaction system used was as follows: the total system was 20 microliters (μl), including 10-100 ng of genomic DNA, 1.0 μl of upstream primers (10 pmol / μl), 1.0 μl of downstream primers (10 pmol / μl), 2.0 μl of 10 ×PCR Buffer (containing magnesium ions Mg 2+ ), 2 μl of dNTPs (0.25 mM), 2.0 U of Taq DNA polymerase and sterilized water.
[0036] The PCR reaction program used was as follows: pre-denaturation at 94°C for 4 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s; ...
Embodiment 3
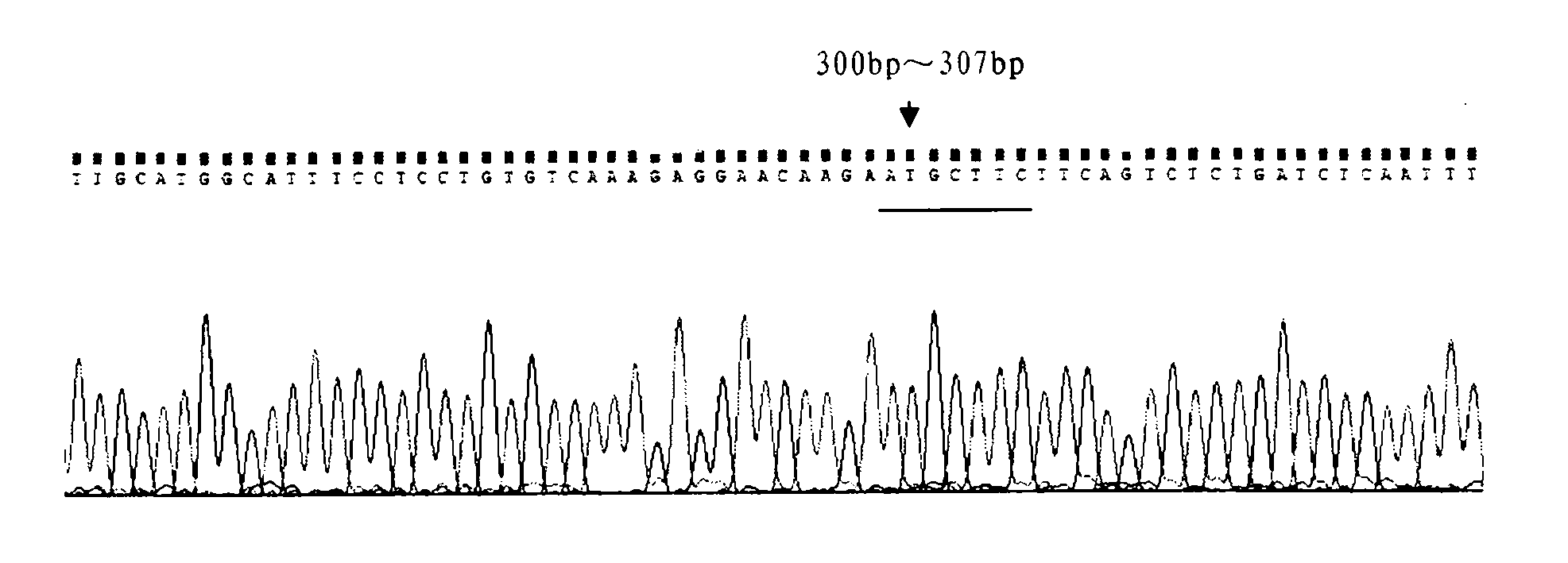
[0039] Genomic DNA was extracted from the blood of the third target bovine by phenol-chloroform extraction, and specific primers were designed to amplify the TLR2 gene promoter region fragment. The primer sequences used were:
[0040] The upstream primer sequence is: SEQ ID NO.4:
[0041] TLR2-promoter-F: 5'-ggattctccaggcaagaaca-3';
[0042] The downstream primer sequence is: SEQ ID NO.5:
[0043] TLR2-promoter-R: 5'-agtcctgtggtggtcccttt-3'.
[0044] The PCR reaction system used was as follows: the total system was 20 microliters (μl), including 10-100 ng of genomic DNA, 1.0 μl of upstream primers (10 pmol / μl), 1.0 μl of downstream primers (10 pmol / μl), 2.0 μl of 10 ×PCR Buffer (containing magnesium ions Mg 2+ ), 2 μl of dNTPs (0.25 mM), 2.0 U of Taq DNA polymerase and sterilized water.
[0045] The PCR reaction program used was as follows: pre-denaturation at 94°C for 4 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com