Methods of identifying genes involved in memory formation using small interfering rna (siRNA)
A memory and gene technology, applied in the field of using small interfering RNA (siRNA) to identify genes involved in memory formation, which can solve the problems of unproven specific role of RNAi, low efficacy of naked siRNA, and limitation of successful siRNA delivery.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
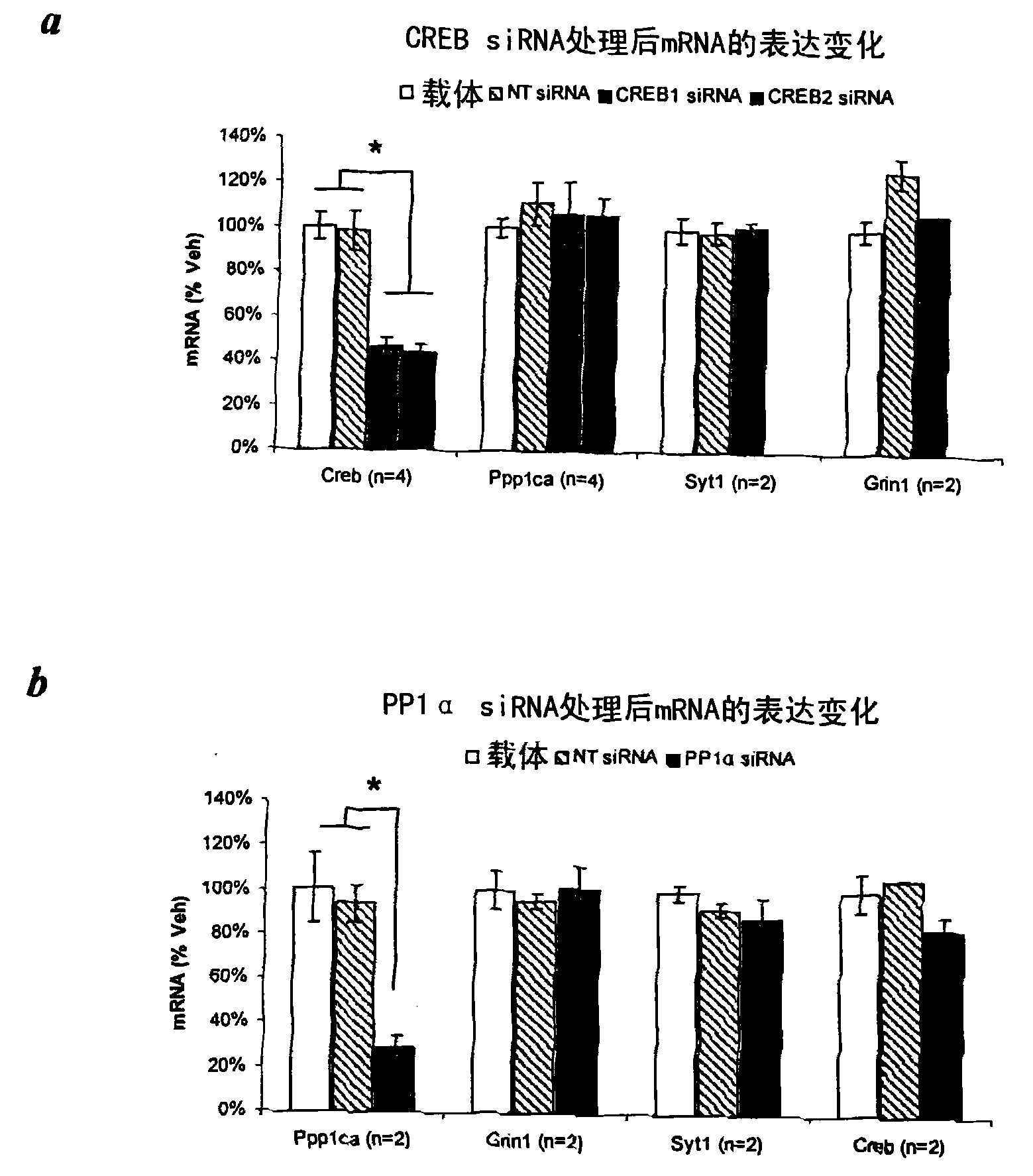
[0115] Example 1 - Screening of siRNA targeting CREB and PPI using Neuro 2A cells
[0116] Screening of a panel of siRNAs targeting the α-isoform of CREB and PP1 in the Neuro2A mouse neuroblastoma cell line. Identify some suitable siRNAs that can efficiently target CREB and PP1α without affecting the mRNA levels of some control genes ( figure 1 ).
[0117] In vivo grade siSTABLE siRNA (Dharmacon Inc., Lafayette, USA). siRNAs are chemically modified to enhance stability. 21-mer siSTABLE non-targeting siRNA was used as a control (sense strand: 5'-UAGCGACUAAACACAUCAAUU-3' (SEQ ID NO: 1); antisense strand: 5'-UUGAUGUGUUUAGUCGCUAUU-3') (SEQ ID NO: 2) (Dharmacon Inc., Lafayette, USA). siRNAs were designed using a multi component rational design algorithm (Reynolds, A. et al. Nat Biotechnol 22, 326-30 (2004)).
[0118] Real-time PCR . Neuro 2A cells were treated with 100 nM siSTABLE siRNA and Dharmafect3 vector (Dharmacon). RNA was isolated using the QIAgen RNeasy kit (Qi...
Embodiment 2
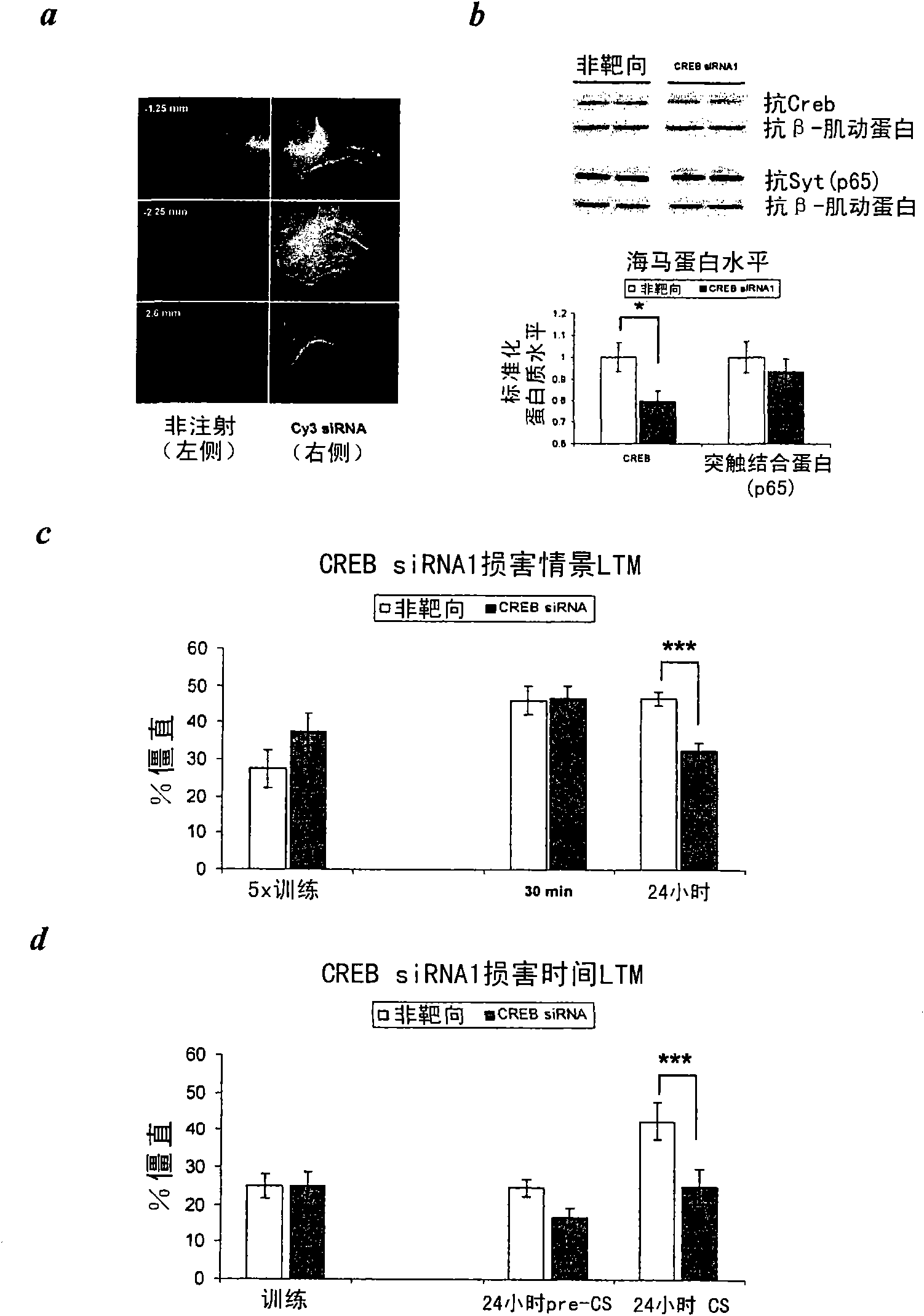
[0131] Example 2 - In vivo delivery of synthetic CREB siRNA in mice
[0132] In vivo delivery of synthetic siRNAs in the CNS is hampered by limited diffusion and uptake.
[0133] Subjects Young adult (10-12 weeks old) C57BL / 6 male mice were used. Upon arrival, mice were housed in groups (5 mice) in standard laboratory cages maintained on a 12:12 hour light-dark cycle. Always perform experiments during the photoperiod. After hippocampal cannulation, mice were housed individually in individual cages and maintained until the end of the experiment. Mice had free access to food and water except for training and testing times. Mice were maintained and bred under standard conditions consistent with the guidelines of the National Institutes of Health (NIH) and approved by the Institution Animal Care and Use Committee.
[0134] Animal surgery and siRNA injection. For siRNA injection, mice were anesthetized with 20 mg / kg Avertin and a 33-gauge catheter (coordinates: A=-1.8 mm, L=+...
Embodiment 3
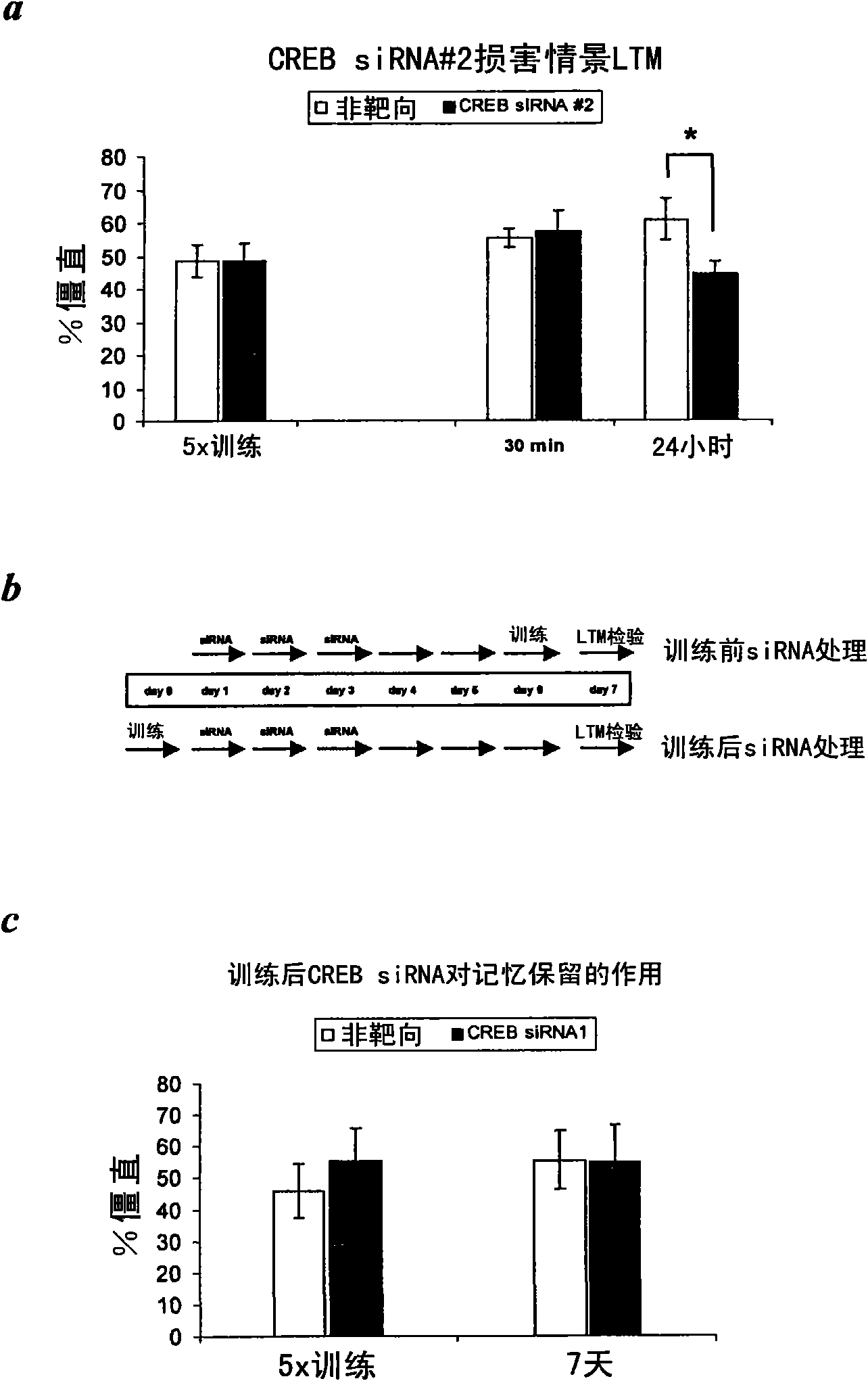
[0142] Example 3 - Effect of siRNA-mediated CREB knockdown on context and trace conditioning
[0143] To examine the effect of siRNA-mediated CREB knockdown on contextual fear conditioning. siRNA targeting the region common to all splice variants of the CREB gene (positions 1114-1132 of NM_009952, corresponding to exon 7 of the CREB gene) was used. Named according to Lonze and Ginty, 2002, Neuron, 35,: 605-623. Mice were treated with CREB siRNA1 or non-targeting control siRNA once a day for 3 consecutive days. Behavioral testing begins after 3 days (see also image 3 b). This design was chosen based on pilot experiments on siRNA knockdown in the hippocampus, as previous studies have shown that gene knockdown by siRNA duplexes occurs in the CNS after a few days ((Salahpour et al., 2007, Biol. Psychiatry 61: 65-69) Tan et al., 2005, Gene Therapy 12: 59-66; Thakker et al., 2004, Proc. Natl. Acad. Sci USA 101: 17270-17275).
[0144]Situational conditioning was performed ess...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com