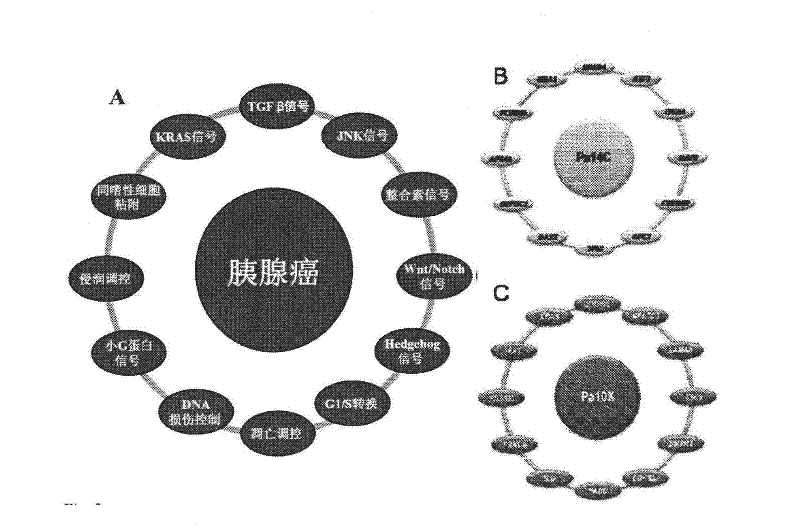
Pathways underlying pancreatic tumorigenesis and an hereditary pancreatic cancer gene
A pancreatic cancer, gene technology, applied in the field of pancreatic cancer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Sample selection. As with all cancer genomics studies, sample selection is critical. For the present study, we selected 24 advanced adenocarcinomas, each from a different and unrelated patient (Table S1). Advanced pancreatic cancers were chosen because they are expected to contain all of the genetic alterations that lead to tumor initiation and progression, whereas early cancers may contain only a subset. 24 cancers were passaged in vitro as cell lines or xenografts in nude mice to facilitate detection of mutations. Studies have shown that this passaging method provides a better DNA template for Sanger sequencing or copy number analysis than the primary tumor because it removes contaminating non-cancerous cells originally present in the tumor (12). Studies have also shown that clonal mutations in cell lines and xenografts are rare, if any, during ex vivo culture (12-14).
Embodiment 2
[0047] Sequencing protocol. Exon sequences encoding proteins found in Conserved Coding Sequences (Release No. 1), Reference Sequences (Release No. 16) and Ensembl Database (Release No. 31) were extracted and used to design primers for genomic DNA amplification (Fig. .S1). The same primers were used if primers designed in our past breast and colorectal cancer studies proved successful (75, 16). New primer sets were designed for 11,579 exons that had not been previously studied and for those exons for which primers had previously been designed but proved suboptimal (see below) (17). Each of the resulting exons was sequenced in 24 pancreatic cancers using dye-terminated sequencing and the 416,622 primers listed in Table S2. Exons containing variant sequences were reamplified from tumor DNA and resequenced to confirm the observed alterations. Additional testing was performed on DNA from normal tissue from patients with mutations in each case. This method determines whether the...
Embodiment 3
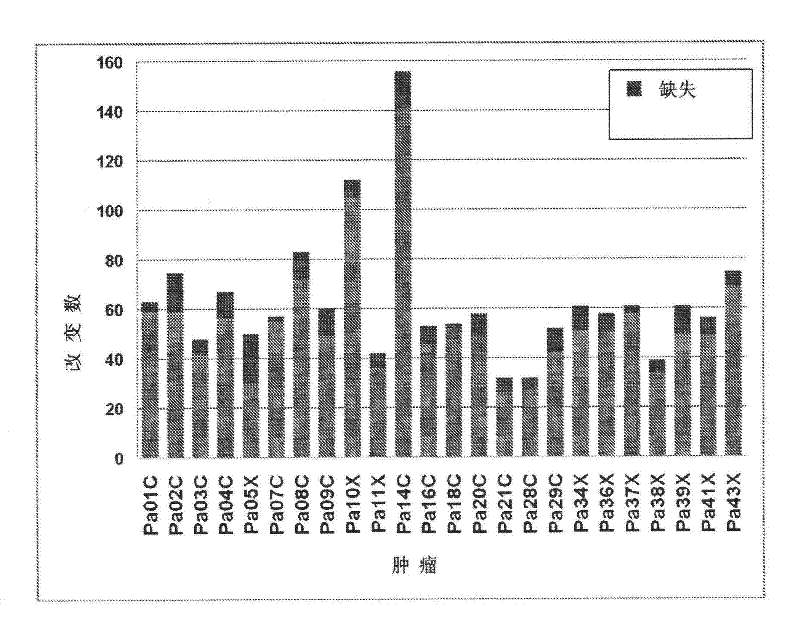
[0050] Somatic mutation. Among the 1562 somatic mutations, 25.5% were synonymous (synonymous), 62.4% were missense (missense), 3.8% were nonsense (nonsense), 5.0% were small insertions and deletions, and 3.3% were in Splice sites (splice sites) or in the untranslated region (UTR) (Table 1). Somatic mutation profiles can provide insights into potential carcinogens and other environmental exposures. Table 1 presents the mutational spectrum observed after a large-scale sequencing analysis of most of the protein-coding genes in the four tumors. Clearly, breast cancer has a unique somatic mutation spectrum, with 5′-TpC mutations predominating and 5′-CpG mutations relatively rare. However, the mutational profiles of colorectal, brain (18) and pancreatic tumors are similar, suggesting that mammary epithelial cells are exposed to different levels or classes of carcinogens compared with cells that develop into other tumors (19, 20) , or use a different repair system. Given that gas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com