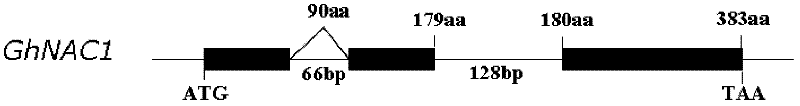Cloning and identification of cotton fiber cell secondary wall development-associated GhNAC1 gene
A gene and gene coding technology, applied in genetic engineering, plant genetic improvement, fermentation, etc., can solve problems such as affecting the assembly of wall components and affecting content.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
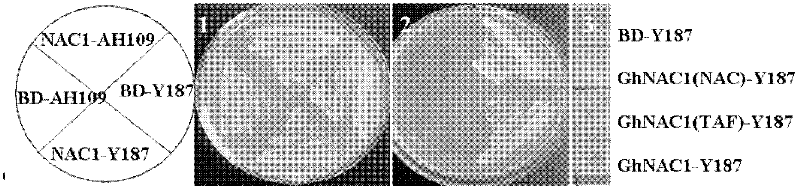
[0033] Cloning identification and functional analysis of the full-length sequence of a new cotton NAC family gene GhANC1:
[0034] 1. Isolation and identification of the full-length sequence of GhNAC1 and its cDNA sequence
[0035] More than 10,000 cotton cDNA clones were isolated from the cotton fiber cDNA library for DNA sequencing. Through bioinformatics analysis, the full-length cDNA sequences of 10 GhNAC genes were obtained, one of which was named GhNAC1. Primers GhNAC1 OEup and GhNAC1 OEdn were designed according to the sequence of GhNAC1 cDNA, and the full-length DNA sequence of the gene was amplified by PCR technology using cotton genome DNA as a template.
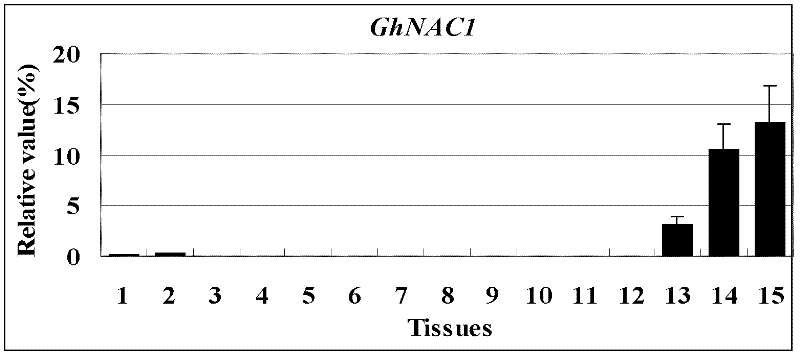
[0036] 2. Quantitative RT-PCR analysis of GhNAC1 gene expression
[0037] (1) Extraction of total RNA from cotton tissue (according to Li XB, Cai L, Cheng NH, Liu JW, 2002. Molecular characterization of the cotton GhTUB1 gene that preferentially expressed in fiber. Plant Physiol. 130: 666-674).
[0038] (2) Real...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com