Novel amplification method and application of ligase reaction mediate
A ligase reaction, mediated technology, applied in the field of amplification, can solve the problems of false positives and limited sensitivity, and achieve the effects of high specificity, efficient amplification and detection, easy design and chemical synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
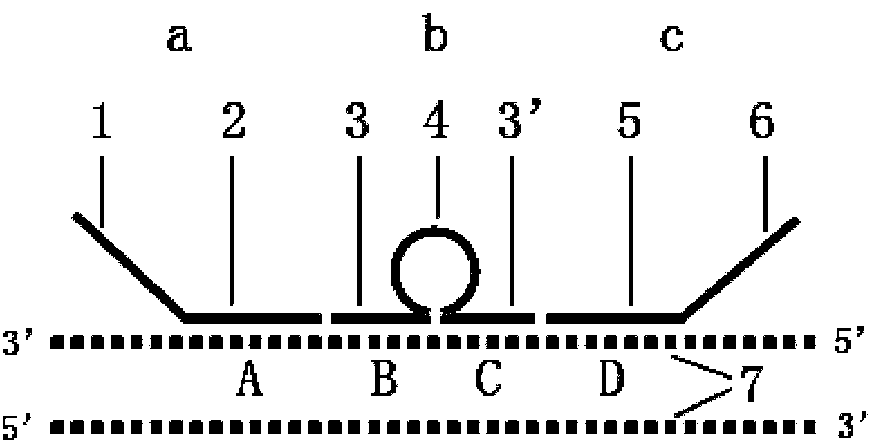
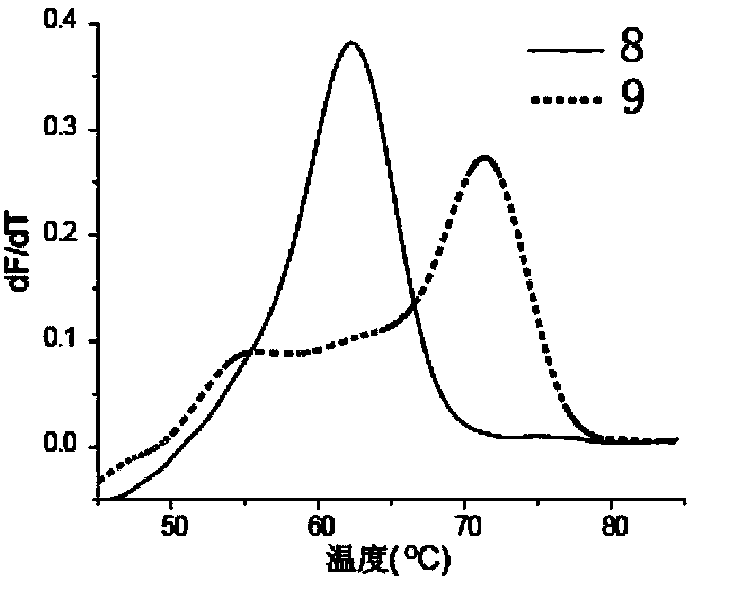
[0032] Embodiment 1: Comparison of melting curves of multiple connection probes:
[0033] target sequence: CTGCAGGGAAATG TCTAGATTGGATCTTGC (SEQ ID NO: 1)
[0034] Ligation probe 1 (hybridizes fully complementary to the target sequence):
[0035] GCAAGATCCAATCTAGACATTTCCCTGCAG (SEQ ID NO: 2)
[0036] Ligation probe 2 (hybridizes with the target sequence to form a hybrid community with a capsule structure, the capsule structure is located in the middle, and the bases at the dotted line form the capsule structure):
[0037] GCAAGATCCAATCTA GGAATCTGGATTCAAAATCTTGACATTTCCCTGCAG
[0038] (SEQ ID NO:3)
[0039] Ligation probe 3 (hybridizes with the target sequence to form a hybrid community of a capsule structure, and the bases at the dotted line position form a capsule structure):
[0040] GCAAGATCCAATCTAGACATTT GGAATCTGGATTCAAAATCTTCCCTGCAG
[0041] (SEQ ID NO:4)
[0042] Ligation probe 4 (hybridizes with the target sequence to form a hybrid community of a capsule struc...
Embodiment 2
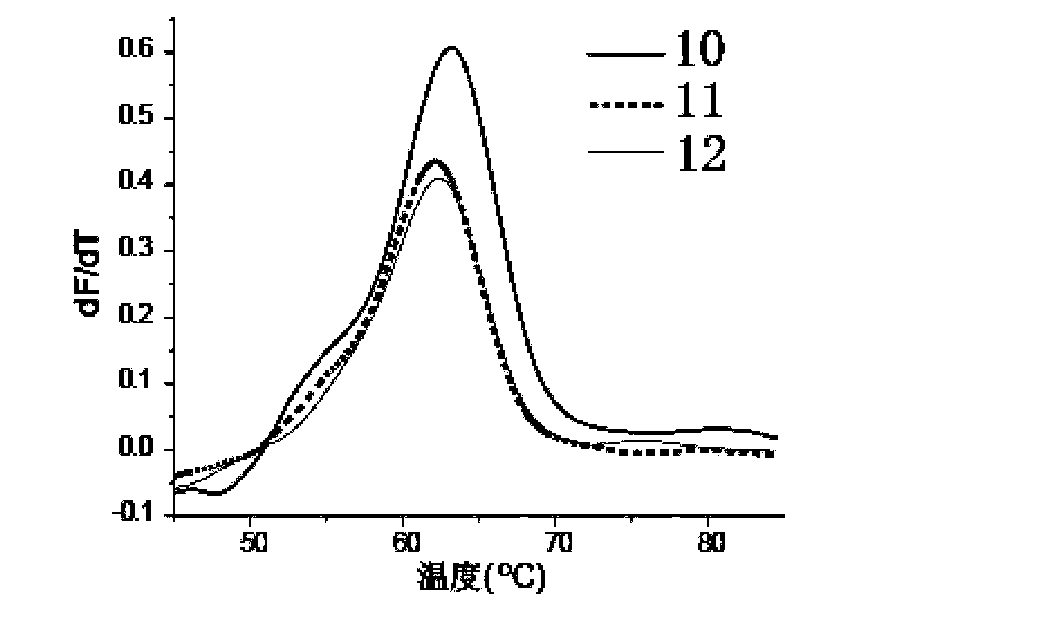
[0049] Embodiment 2: Quantitative detection target sequence
[0050] Select an artificially synthesized DNA sequence as the object of investigation, and design three ligation probes for this sequence to hybridize to the positions adjacent to the target sequence. The specific sequence is as follows:
[0051] target sequence:
[0052] CTACACAGTCTCCTGTACCTGGGCAATATGATGCTACCAAATTTAAGCAGTATAGCAGACATGTTGAGGAATATGA (SEQ ID NO: 7)
[0053] Connect probe a:
[0054] TGGAGCGACGATACGAAGATA TCATATTCCTCAACATGTCTGC (SEQ ID NO: 8)
[0055] Connect probe b:
[0056] PO4-TATACTGCTTAAATTT AACTTCGGTCCTTCATCGCT GGTAGCATCATATTGC
[0057] (SEQ ID NO:9)
[0058] Connect probe c:
[0059] PO4-CCAGGTACAGGAGACTGTGTAG TCTAGATAGGATCTTGGAGC
[0060] (SEQ ID NO: 10)
[0061] Wherein, the upstream primer tag sequence of connection probe a is the universal primer F sequence, and the downstream primer binding tag sequence of connectio...
Embodiment 3
[0068] Example 3: SNP Genotyping Detection of Three Human Genomic DNA Samples
[0069] Select the SNP site (rs740598) as the research object, and design four connection probes as follows:
[0070] Connect probe a:
[0071] TGGAGCGACGATACGAAGATA CCAAATATTTTTCGTAAGTATTTCAAAT
[0072] (SEQ ID NO: 14)
[0073]To connect probe b-1:
[0074] PO4-AGCAATGGCTCGTC CATCTCTAAGGCAAGGCTC TATGGTTAGTCTCA
[0075] (SEQ ID NO: 15)
[0076] To connect probe b-2:
[0077] PO4-AGCAATGGCTCGTC ACCTTCCGTCTGTACTCGT TATGGTTAGTCTCG
[0078] (SEQ ID NO: 16)
[0079] Connect probe c:
[0080] PO4-CAGCCACATTCTCAGAACTGC TCTAGATAGGATCTTGGAGC (SEQ ID NO: 17)
[0081] Wherein, the tag sequences of connecting probes a and c are the same as in Example 2, and the detection tag sequence of connecting probe b-1 is a FAM-labeled Taqman probe sequence (FAM-CATCTCTAAGGCAAGGCTC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com