Polymerase chain reaction (PCR) method for specifically detecting pectobacterium carotovorum
A technology of carrot soft rot and pectobacillus, which is applied in the directions of microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve problems such as poor bacterial specificity, and achieve high specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment one detects diseased plant
[0033] Taking artichoke rhizomes as an example, Pectinbacterium carotovora soft rot, which causes artichoke root rot disease, was detected.
[0034] Inoculate the strains isolated from artichoke rhizome rot diseased plants and identified as Pectinobacillus carotovora soft rot on disease-free artichoke stems, and after the initial symptoms appear, extract DNA from the diseased tissue as a PCR template, and then use the present invention to The primer pair was used for PCR, and the PCR parameters were the same as before. PCR results showed that artichoke stems inoculated with P. carotovora were positive, while disease-free artichoke stems not inoculated with P. carotovora were negative, see Figure 5 . It shows that the specific primer can be used to detect the diseases caused by Pectinbacterium carotovora soft rot.
Embodiment 2
[0035] Embodiment two distinguishes similar bacteria
[0036] According to the aforementioned bacterial DNA extraction and PCR methods, 4 kinds of bacteria were tested: Pectinobacillus carotovora, Blackleg of potato, Erchetia chrysanthemi and Pantoea pineapple. Only the sample of Pectinobacillus carotovora was positive, and the other three were positive. Bacterial sample was negative, see Figure 6 .
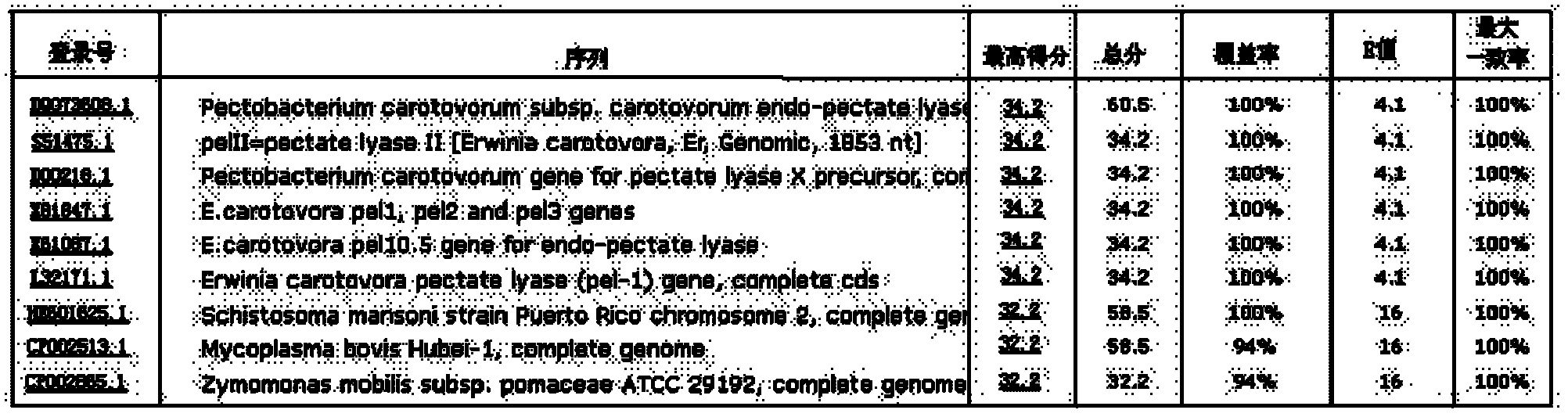
[0037] Thus, theoretically, the results of BLAST of the L32171.1 sequence and forward and reverse primers, and practically, the results of Examples 1 and 2 all confirm the specificity of the primer pair of the present invention. Using this pair of primers for PCR detection can accurately distinguish Pectinobacillus carotovora from other bacteria, and can distinguish Pectinobacillus carotovora from other bacteria of the genus Pectinbacterium.
[0038]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com