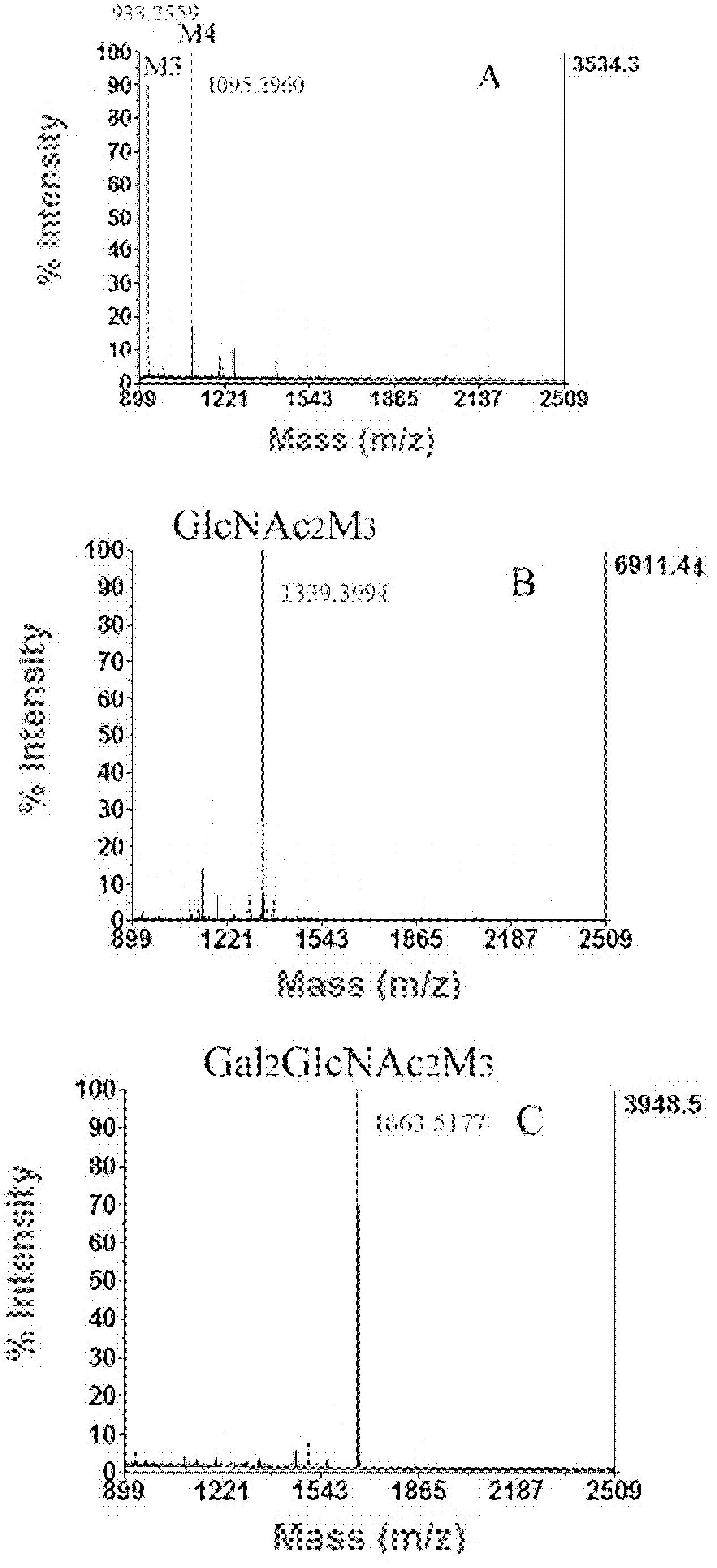Humanized glycosylation modified hansenula polymorpha
A Hansenula yeast, glycosylated protein technology, applied in microorganism-based methods, biochemical equipment and methods, introduction of foreign genetic material using vectors, etc., can solve the problem of Hansenula's human-derived glycosylation transformation has not yet been successful. cases, etc., to achieve the effect of eliminating antigenicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Embodiment 1, the construction of recombinant plasmid A and engineering bacteria A
[0051] 1. Construction of recombinant plasmid A
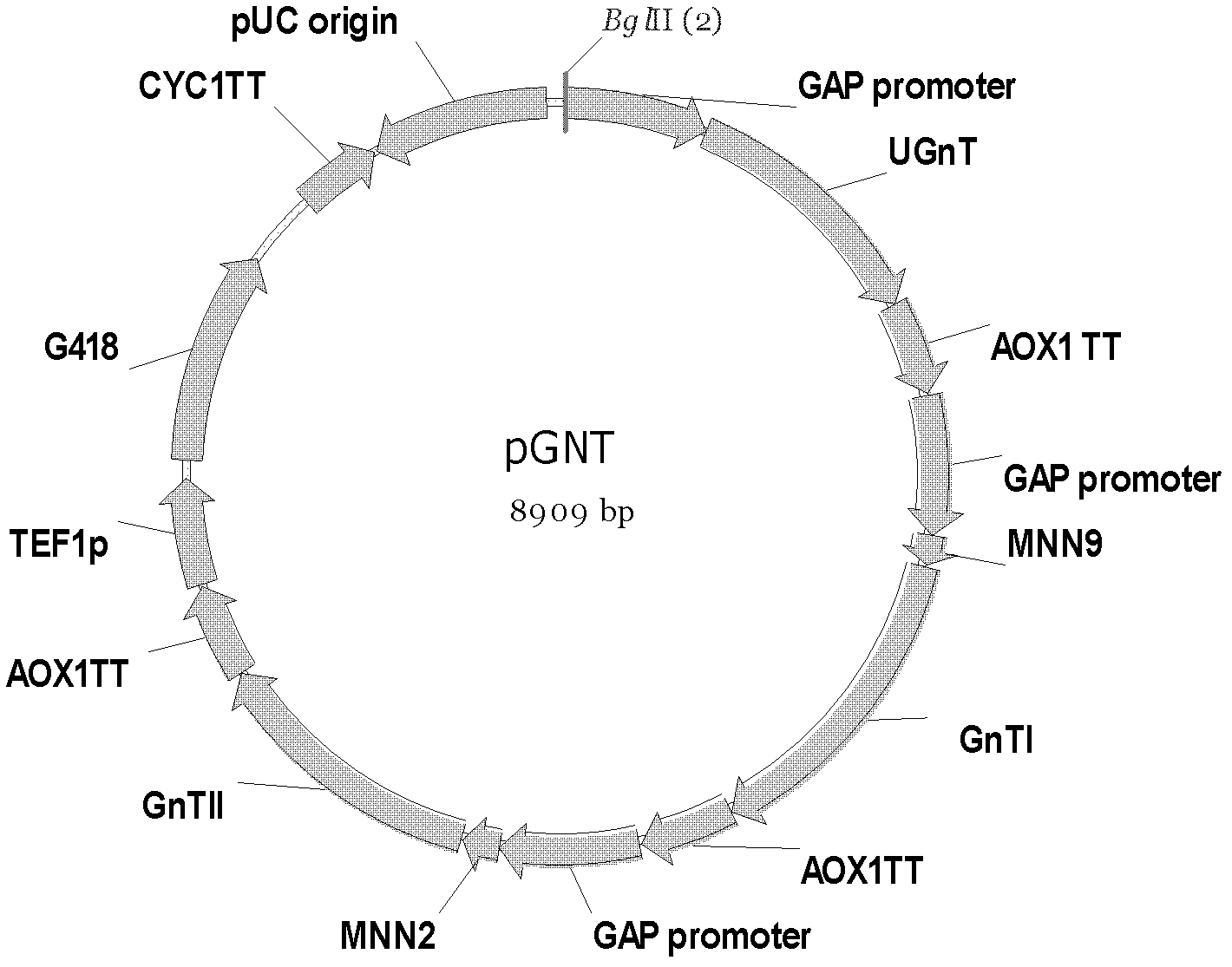
[0052] The schematic diagram of the structure of recombinant plasmid pGNT (also known as recombinant plasmid A) is shown in figure 1 . The nucleotide sequence of the recombinant plasmid pGNT is shown in sequence 1 of the sequence listing. In sequence 1, the 1st to 6th nucleotides from the 5' end are the BglII restriction recognition sequence, the 7th to 541st nucleotides are the GAP promoter, the 548th to 1535th nucleotides are the UGnT gene, and the 1559th nucleotide The 1896th nucleotide is the AOX1TT termination sequence, the 1903rd to 2437th nucleotide is the GAP promoter, the 2438th to 2551st nucleotide is the MMN9 gene, the 2558th to 3781st nucleotide is the GnTI gene, the 3806th nucleotide Nucleotides 4143 to 4143 are AOX1TT termination sequences, nucleotides 4150 to 4684 are GAP promoters, nucleotides 4685 to 4825 are MNN2 gen...
Embodiment 2
[0107] Embodiment 2, the construction of recombinant plasmid B and engineering bacteria B
[0108] 1. Construction of recombinant plasmid B
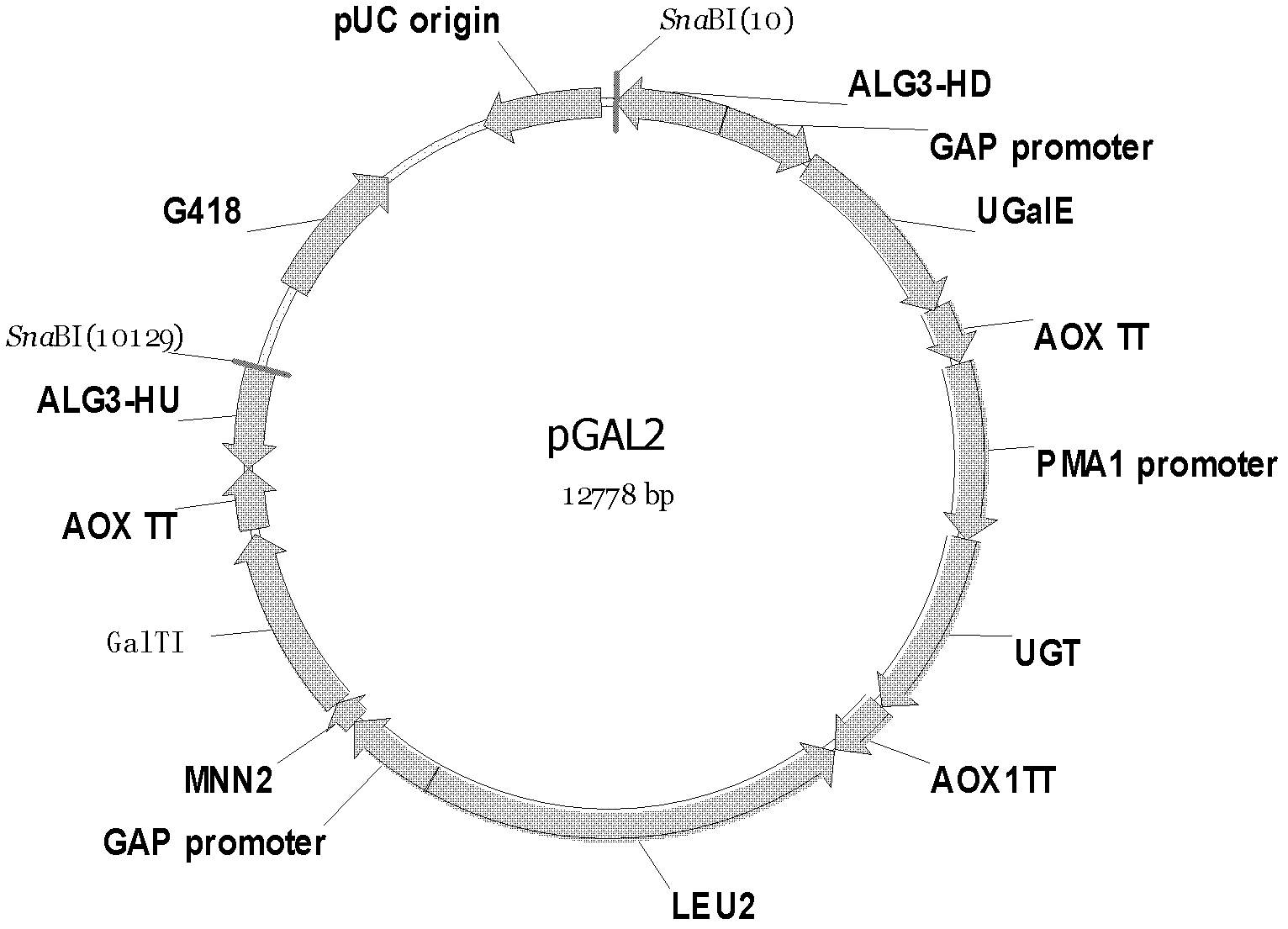
[0109] The schematic diagram of the structure of recombinant plasmid PGAL2 (also known as recombinant plasmid B) is shown in figure 2 . The nucleotide sequence of the recombinant plasmid PGAL2 is shown in sequence 2 of the sequence listing. In sequence 2, the 6th to 11th nucleotides from the 5' end are the SnaBI restriction recognition sequence, the 12th to 616th nucleotides are the ALG3-HD fragment (homologous arm), and the 623rd to 1157th nucleotides It is the GAP promoter, the 1164th to 2231st nucleotide is the UGalE gene, the 2256th to 2593rd nucleotide is the AOX1TT termination sequence, the 2600th to 3600th nucleotide is the PMA1 promoter, the 3601st to 4659th nucleotide The acid is the UGT gene, the 4684th to 5021st nucleotide is the AOX1TT termination sequence, the 5028th to 7433rd nucleotide is the LEU2 gene, the 7440th to 7...
Embodiment 3
[0171] Embodiment 3, purification and identification of glycoprotein expressed by engineering bacteria
[0172] 1. Introduce the plasmid pHFMDZαL2-rGOD into the engineering strain A to obtain the recombinant strain H.pGNT-rGOD.
[0173] 2. The plasmid pHFMDZαL2-rGOD was introduced into the engineering strain B to obtain the recombinant strain H.pGAL-rGOD.
[0174] 3. The plasmid pHFMDZαL2-rGOD was introduced into Hansenula spp. Hpalg3Δalg11ΔpRft1 to obtain control bacteria.
[0175] 4. The recombinant bacteria H.pGNT-rGOD obtained in step 1, the recombinant bacteria H.pGAL-rGOD obtained in step 2, and the control bacteria obtained in step 3 were subjected to the following experiments:
[0176] (1) Induced expression of rGOD protein
[0177] ① Inoculate a single colony of engineering bacteria into a test tube containing 5mL SD medium, and incubate at 30°C for 48h; then transfer it to a 250mL Erlenmeyer flask containing 50mL YNG medium, and incubate at 30°C for 32 hours (30-35...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com